5WJH
 
 | | Using sound pulses to solve the crystal harvesting bottleneck | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CALCIUM ION, Proteinase K | | Authors: | Soares, A.S, Brennan, H.M, McCarthy, L, Leroy, L. | | Deposit date: | 2017-07-22 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Using sound pulses to solve the crystal-harvesting bottleneck.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5VGJ
 
 | | Crystal Structure of the Human Fab VRC38.01, an HIV-1 V1V2-Directed Neutralizing Antibody Isolated from Donor N90, bound to a scaffolded WITO V1V2 domain | | Descriptor: | 1FD6-V1V2-WITO, 2-acetamido-2-deoxy-beta-D-glucopyranose, VRC38.01 Fab Heavy Chain, ... | | Authors: | Gorman, J, Li, J, Kwong, P.D. | | Deposit date: | 2017-04-11 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.456 Å) | | Cite: | Virus-like Particles Identify an HIV V1V2 Apex-Binding Neutralizing Antibody that Lacks a Protruding Loop.
Immunity, 46, 2017
|
|
5W8E
 
 | | The structure of a CoA-dependent acyl-homoserine lactone synthase, BjaI, with the adduct of SAH and IV-CoA | | Descriptor: | (2S)-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)-2-[(3-methylbutanoyl)amino]butanoic acid, ADENINE, Autoinducer synthase, ... | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2017-06-21 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the substrate specificity of quorum signal synthases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7X05
 
 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with the nascent chitooligosaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin synthase, MANGANESE (II) ION, ... | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7X06
 
 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with UDP | | Descriptor: | Chitin synthase, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
6BQK
 
 | |
7WJM
 
 | |
7WJN
 
 | | CryoEM structure of chitin synthase 1 mutant E495A from Phytophthora sojae complexed with UDP-GlcNAc | | Descriptor: | Chitin synthase, MANGANESE (II) ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7WJO
 
 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with nikkomycin Z | | Descriptor: | (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
6BQJ
 
 | |
5ENM
 
 | | Compound 10 | | Descriptor: | (2~{R},4~{S},6~{S})-4-[2,4-bis(fluoranyl)-5-pyrimidin-5-yl-phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-1,3-thiazinan-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Lewis, H.A. | | Deposit date: | 2015-11-09 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Targeting the BACE1 Active Site Flap Leads to a Potent Inhibitor That Elicits Robust Brain A beta Reduction in Rodents.
Acs Med.Chem.Lett., 7, 2016
|
|
5ENK
 
 | | Compound 18 | | Descriptor: | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)-5-pyrimidin-5-yl-phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Lewis, H.A. | | Deposit date: | 2015-11-09 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Targeting the BACE1 Active Site Flap Leads to a Potent Inhibitor That Elicits Robust Brain A beta Reduction in Rodents.
Acs Med.Chem.Lett., 7, 2016
|
|
3PHE
 
 | | HCV NS5B with a bound quinolone inhibitor | | Descriptor: | 4-chlorobenzyl 6-fluoro-7-(4-methylpiperazin-1-yl)-1-[4-(methylsulfonyl)benzyl]-4-oxo-1,4-dihydroquinoline-3-carboxylate, HCV encoded nonstructural 5B protein | | Authors: | Somoza, J.R, To, N, Lehoux, I. | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Quinolones as HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5EWI
 
 | |
6UGJ
 
 | |
6UGG
 
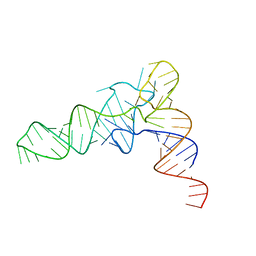 | | Structure of unmodified E. coli tRNA(Asp) | | Descriptor: | tRNAasp | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2019-09-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of an unmodified bacterial tRNA reveal intrinsic structural flexibility and plasticity as general properties of unbound tRNAs.
Rna, 26, 2020
|
|
7RJ8
 
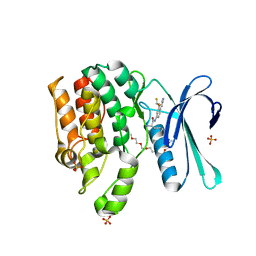 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 ISOFORM 1 COMPLEXED WITH LIGAND (2R)-2-AMINO-N-[3-(DIFLUOROM ETHOXY)-4-(1,3-OXAZOL-5-YL)PHENYL]-4-METHYLPENTANAMIDE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 5-[(4-aminopiperidin-1-yl)methyl]-N-{3-[5-(propan-2-yl)-1,3,4-thiadiazol-2-yl]phenyl}pyrrolo[2,1-f][1,2,4]triazin-4-amine, AP2-associated protein kinase 1, ... | | Authors: | Pokross, M, Muckelbauer, J. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Bicyclic Heterocyclic Replacement of an Aryl Amide Leading to Potent and Kinase-Selective Adaptor Protein 2-Associated Kinase 1 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RJ6
 
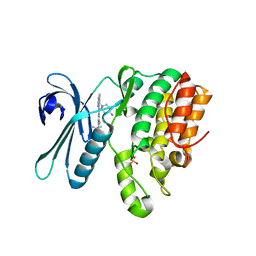 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 ISOFORM 1 COMPLEXED WITH LIGAND (2R)-2-AMINO-N-[3-(DIFLUOROM ETHOXY)-4-(1,3-OXAZOL-5-YL)PHENYL]-4-METHYLPENTANAMIDE | | Descriptor: | (1S)-1-[4-ethyl-6-(1,3-oxazol-5-yl)quinazolin-2-yl]-3-methylbutan-1-amine, 5-[(4-aminopiperidin-1-yl)methyl]-N-{3-[5-(propan-2-yl)-1,3,4-thiadiazol-2-yl]phenyl}pyrrolo[2,1-f][1,2,4]triazin-4-amine, AP2-associated protein kinase 1, ... | | Authors: | Muckelbauer, J. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.132 Å) | | Cite: | Bicyclic Heterocyclic Replacement of an Aryl Amide Leading to Potent and Kinase-Selective Adaptor Protein 2-Associated Kinase 1 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RJ7
 
 | |
6UGI
 
 | |
7KCF
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-24512 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-hydroxyphenyl)-5-methyl-2-phenyl-3-(piperidin-1-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one, GLYCEROL, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7KDA
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor compound 34 | | Descriptor: | 2,3-diphenyl-5-[(1H-pyrazol-3-yl)amino]pyrazolo[1,5-a]pyrimidin-7(4H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7KDB
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor compound 35 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(4-hydroxyphenyl)-2,3-diphenyl-5-[(1H-pyrazol-3-yl)amino]pyrazolo[1,5-a]pyrimidin-7(4H)-one, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7KCE
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor compound 2 | | Descriptor: | 5-methyl-2,3-diphenylpyrazolo[1,5-a]pyrimidin-7(4H)-one, CHLORIDE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7KCC
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AG-270 | | Descriptor: | 1,2-ETHANEDIOL, 3-(cyclohex-1-en-1-yl)-6-(4-methoxyphenyl)-2-phenyl-5-[(pyridin-2-yl)amino]pyrazolo[1,5-a]pyrimidin-7(4H)-one, CHLORIDE ION, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
