2O1B
 
 | | Structure of aminotransferase from Staphylococcus aureus | | Descriptor: | Aminotransferase, class I, PYRIDOXAL-5'-PHOSPHATE | | Authors: | McGrath, T.E, Dharamsi, A, Thambipillai, D, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of aminotransferase from Staphylococcus aureus
To be Published
|
|
2IP4
 
 | | Crystal Structure of Glycinamide Ribonucleotide Synthetase from Thermus thermophilus HB8 | | Descriptor: | Phosphoribosylamine--glycine ligase, SULFATE ION | | Authors: | Sampei, G, Baba, S, Kanagawa, M, Yanai, H, Ishii, T, Kawai, H, Fukai, Y, Ebihara, A, Nakagawa, N, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-11 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2EJ5
 
 | | Crystal structure of GK2038 protein (enoyl-CoA hydratase subunit II) from Geobacillus kaustophilus | | Descriptor: | Enoyl-CoA hydratase subunit II | | Authors: | Okazaki, N, Agari, Y, Ebihara, A, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GK2038 protein (enoyl-CoA hydratase subunit II) from Geobacillus kaustophilus
To be Published
|
|
2ECR
 
 | | Crystal structure of the ligand-free form of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-13 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2EGZ
 
 | | Crystal structure of the 3-dehydroquinate dehydratase from Aquifex aeolicus VF5 | | Descriptor: | 3-dehydroquinate dehydratase, L(+)-TARTARIC ACID | | Authors: | Karthe, P, Kumarevel, T.S, Ebihara, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-02 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the 3-dehydroquinate dehydratase from Aquifex aeolicus VF5
To be Published
|
|
2ED4
 
 | | Crystal structure of flavin reductase HpaC complexed with FAD and NAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygenae | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2EHD
 
 | | Crystal Structure Analysis of Oxidoreductase | | Descriptor: | COBALT (II) ION, Oxidoreductase, short-chain dehydrogenase/reductase family | | Authors: | Kamiya, N, Hikima, T, Ebihara, A, Inoue, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-06 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of putative oxidoreductase from Thermus thermophilus HB8
to be published
|
|
2EIS
 
 | | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8 | | Descriptor: | COENZYME A, Hypothetical protein TTHB207 | | Authors: | Kamitori, S, Yoshida, H, Satoh, S, Iino, H, Ebihara, A, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8
To be Published
|
|
2ECU
 
 | | Crystal structure of flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, DODECAETHYLENE GLYCOL, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygnease | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2E6K
 
 | | X-ray structure of Thermus thermopilus HB8 TT0505 | | Descriptor: | Transketolase | | Authors: | Yoshida, H, Kamitori, S, Agari, Y, Iino, H, Kanagawa, M, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-27 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-ray structure of Thermus thermophilus HB8 TT0505
To be Published
|
|
2HIQ
 
 | | Crystal structure of JW1657 from Escherichia coli | | Descriptor: | Hypothetical protein ydhR | | Authors: | Chen, L.Q, Chen, L.R, Liu, Z.-J, Temple, W, Lee, D, Chang, S.-H, Rose, J.P, Ebihara, A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-06-29 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of JW1657 from Escherichia coli at 2.0A resolution
To be Published
|
|
3K8U
 
 | | Crystal Structure of the Peptidase Domain of Streptococcus ComA, a Bi-functional ABC Transporter Involved in Quorum Sensing Pathway | | Descriptor: | Putative ABC transporter, ATP-binding protein ComA | | Authors: | Ishii, S, Yano, T, Ebihara, A, Okamoto, A, Manzoku, M, Hayashi, H. | | Deposit date: | 2009-10-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the peptidase domain of Streptococcus ComA, a bifunctional ATP-binding cassette transporter involved in the quorum-sensing pathway
J.Biol.Chem., 285, 2010
|
|
2IC7
 
 | | Crystal Structure of Maltose Transacetylase from Geobacillus kaustophilus | | Descriptor: | Maltose transacetylase | | Authors: | Liu, Z.J, Li, Y, Chen, L, Zhu, J, Rose, J.P, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-12 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Maltose Transacetylase From Geobacillus kaustophilus at 1.78 Angstrom Resolution
To be Published
|
|
2ICU
 
 | | Crystal Structure of Hypothetical Protein YedK From Escherichia coli | | Descriptor: | Hypothetical protein yedK | | Authors: | Chen, L, Liu, Z.J, Li, Y, Zhao, M, Rose, J, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Hypothetical Protein YedK From Escherichia coli
To be Published
|
|
3AW8
 
 | | Crystal structure of N5-carboxyaminoimidazole ribonucleotide synthetase from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Okada, K, Tsunoda, S, Taka, H, Baba, S, Kanagawa, M, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-03-15 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of N5-carboxyaminoimidazole ribonucleotide synthetase, PurK, from thermophilic bacteria
To be Published
|
|
3CQ2
 
 | | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (other form) from Thermus Thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB138 | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Satoh, S, Kitamura, Y, Ebihara, A, Chen, L, Liu, Z.J, Wang, B.C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein from Thermus Thermophilus HB8
To be Published
|
|
3AX6
 
 | | Crystal structure of N5-carboxyaminoimidazole ribonucleotide synthetase from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoribosylaminoimidazole carboxylase, ATPase subunit | | Authors: | Miyazawa, R, Kanagawa, M, Baba, S, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of N5-carboxyaminoimidazole ribonucleotide synthetase, PurK, from thermophilic bacteria
To be Published
|
|
5XM0
 
 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3.3, and H4 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.874 Å) | | Cite: | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
5XM1
 
 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3mm7, and H4 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
7VQP
 
 | | Vitamin D receptor complexed with a lithocholic acid derivative | | Descriptor: | 3-((R)-4-((3R,5R,8R,9S,10S,13R,14S,17R)-3-(2-hydroxy-2-methylpropyl)-10,13-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-17-yl)pentanamido)propanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, K, Numoto, N, Kagechika, H, Tanatani, A, Ito, N. | | Deposit date: | 2021-10-20 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lithocholic Acid Amides as Potent Vitamin D Receptor Agonists.
Biomolecules, 12, 2022
|
|
1CYD
 
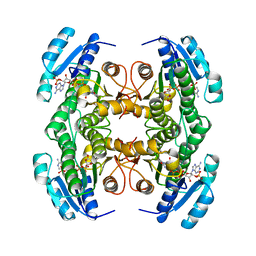 | | CARBONYL REDUCTASE COMPLEXED WITH NADPH AND 2-PROPANOL | | Descriptor: | CARBONYL REDUCTASE, ISOPROPYL ALCOHOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1995-09-01 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ternary complex of mouse lung carbonyl reductase at 1.8 A resolution: the structural origin of coenzyme specificity in the short-chain dehydrogenase/reductase family.
Structure, 4, 1996
|
|
2DQT
 
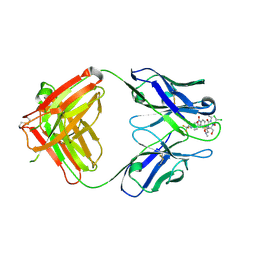 | | High resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
2DQU
 
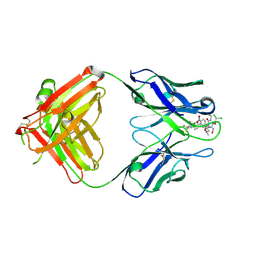 | | Crystal form II: high resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
6KX8
 
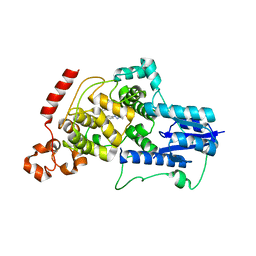 | | Crystal structure of mouse Cryptochrome 2 in complex with TH301 compound | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-2 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
6KX7
 
 | | Crystal structure of mouse Cryptochrome 1 in complex with TH301 compound | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-1 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
