8R8K
 
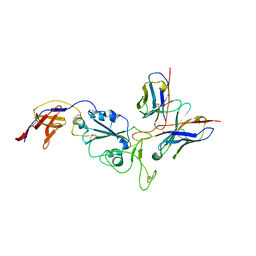 | | XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | Spike glycoprotein,Fibritin, XBB-4 Fab Heavy chain, XBB-4 Fab Light chain | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
7Q9P
 
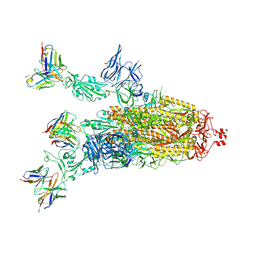 | | Beta-06 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-06 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q9F
 
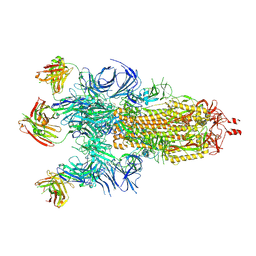 | | Beta-50 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q9J
 
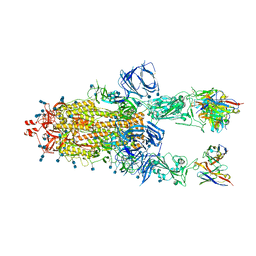 | | Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-26 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q6E
 
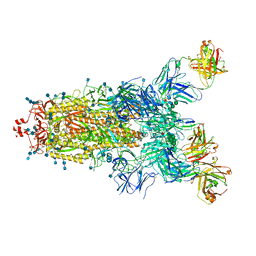 | | Beta049 fab in complex with SARS-CoV2 beta-Spike glycoprotein, The Beta mAb response underscores the antigenic distance to other SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-49 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-07 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q9G
 
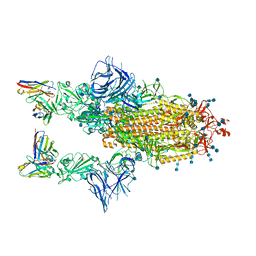 | | COVOX-222 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q9I
 
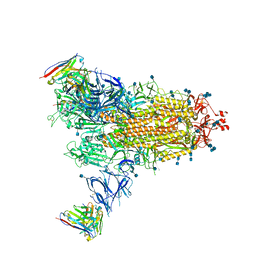 | | Beta-43 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-43 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q9K
 
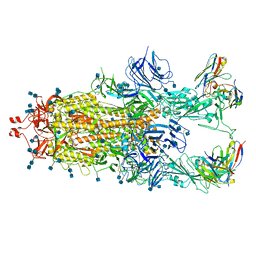 | | Beta-32 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-32 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q9M
 
 | | Beta-53 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-53 fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
6YOR
 
 | | Structure of the SARS-CoV-2 spike S1 protein in complex with CR3022 Fab | | Descriptor: | IgG H chain, IgG L chain, Spike glycoprotein | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Duyvesteyn, H.M.E, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-15 | | Release date: | 2020-04-29 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
8Q3L
 
 | |
8R80
 
 | | SARS-CoV-2 Delta RBD in complex with XBB-9 Fab and an anti-Fab nanobody | | Descriptor: | Spike protein S1, XBB-9 Fab heavy chain, XBB-9 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-27 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.03 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
6U57
 
 | |
6FTR
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wiedorn, M.O, Oberthuer, D, Barty, A, Chapman, H.N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76000106 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
6GTH
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Wiedorn, M, Oberthuer, D, Werner, N, Schubert, R, White, T.A, Mancuso, A, Perbandt, M, Betzel, C, Barty, A, Chapman, H. | | Deposit date: | 2018-06-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
7Q0A
 
 | | SARS-CoV-2 Spike ectodomain with Fab FI3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI3A fab Light chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
7ZR9
 
 | | OMI-2 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-2 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-03 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZRC
 
 | | OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-38 Fab Heavy Chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-04 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZR7
 
 | | OMI-42 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-42 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-03 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZR8
 
 | | OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-38 Fab light chain, Omi-38 fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-03 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
8BCZ
 
 | | SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45 | | Descriptor: | BA.2-23 heavy chain, BA.2-23 light chain, BA.2-36 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
4X4I
 
 | |
4X4E
 
 | |
4X4G
 
 | |
4X4F
 
 | |
