7YP0
 
 | | Crystal structure of CtGST | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2022-08-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8K6Z
 
 | | NMR structure of human leptin | | Descriptor: | Leptin | | Authors: | Fan, X, Qin, R, Yuan, W, Fan, J, Huang, W, Lin, Z. | | Deposit date: | 2023-07-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human leptin reveals a conformational plasticity important for receptor recognition.
Structure, 32, 2024
|
|
8K4S
 
 | | CryoEM structure of Gq coupled MRGPRX4 with agonist DCA-3P | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},12~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-12-oxidanyl-3-phosphonooxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2023-07-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided discovery of bile acid derivatives for treating liver diseases without causing itch
Cell(Cambridge,Mass.), 2024
|
|
8KEX
 
 | | CryoEM structure of Gq coupled MRGPRX4 with agonist DCA-3P, local | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},12~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-12-oxidanyl-3-phosphonooxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Soluble cytochrome b562,Mas-related G-protein coupled receptor member X4,Green fluorescent protein | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2023-08-14 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-guided discovery of bile acid derivatives for treating liver diseases without causing itch
Cell(Cambridge,Mass.), 2024
|
|
2K3Q
 
 | |
2K3P
 
 | |
7JM5
 
 | | Crystal structure of KDM4B in complex with QC6352 | | Descriptor: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, Lysine-specific demethylase 4B, NICKEL (II) ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-07-31 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting KDM4 for treating PAX3-FOXO1-driven alveolar rhabdomyosarcoma.
Sci Transl Med, 14, 2022
|
|
2K3N
 
 | |
2K3O
 
 | |
5VLR
 
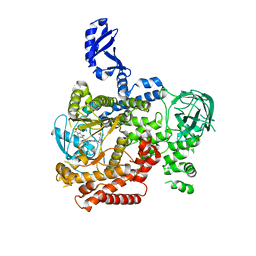 | | CRYSTAL STRUCTURE OF PI3K DELTA IN COMPLEX WITH A TRIFLUORO-ETHYL-PYRAZOL-PYROLOTRIAZINE INHIBITOR | | Descriptor: | 4-acetyl-1-(3-{4-amino-5-[1-(2,2,2-trifluoroethyl)-1H-pyrazol-5-yl]pyrrolo[2,1-f][1,2,4]triazin-7-yl}phenyl)-3,3-dimethylpiperazin-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Sack, J.S. | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a Potent, Selective, and Efficacious Phosphatidylinositol 3-Kinase delta (PI3K delta ) Inhibitor for the Treatment of Immunological Disorders.
J. Med. Chem., 60, 2017
|
|
1M5Z
 
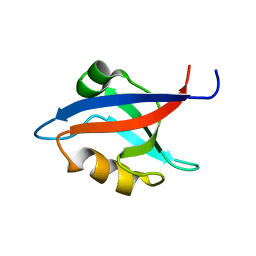 | | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area | | Descriptor: | AMPA receptor interacting protein | | Authors: | Feng, W, Fan, J, Jiang, M, Shi, Y, Zhang, M. | | Deposit date: | 2002-07-11 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area
J.Biol.Chem., 277, 2002
|
|
4X7Q
 
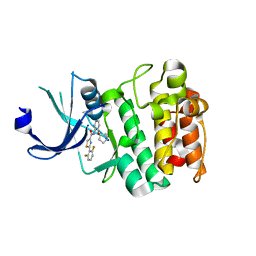 | | PIM2 kinase in complex with Compound 1s | | Descriptor: | 2-(2,6-difluorophenyl)-N-{4-[(3S)-pyrrolidin-3-yloxy]pyridin-3-yl}-1,3-thiazole-4-carboxamide, PHOSPHATE ION, Serine/threonine-protein kinase pim-2 | | Authors: | Marcotte, D.J, Silvian, L.F. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-based design of low-nanomolar PIM kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8DNT
 
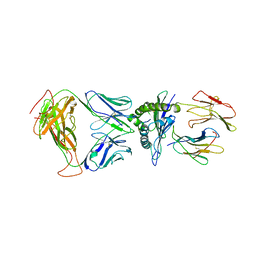 | | SARS-CoV-2 specific T cell receptor | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, Nucleoprotein, ... | | Authors: | Gallagher, D.T, Wu, D, Gowthaman, R, Pierce, B.G, Mariuzza, R.A, Weng, N.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | SARS-CoV-2 infection establishes a stable and age-independent CD8 + T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
Nat Commun, 14, 2023
|
|
1W24
 
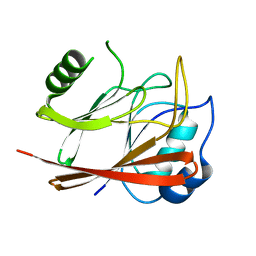 | | Crystal Structure Of human Vps29 | | Descriptor: | VACUOLAR PROTEIN SORTING PROTEIN 29 | | Authors: | Wang, D, Guo, M, Teng, M, Niu, L. | | Deposit date: | 2004-06-26 | | Release date: | 2005-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Vacuolar Protein Sorting Protein 29 Reveals a Phosphodiesterase/Nuclease-Like Fold and Two Protein-Protein Interaction Sites.
J.Biol.Chem., 280, 2005
|
|
2M0M
 
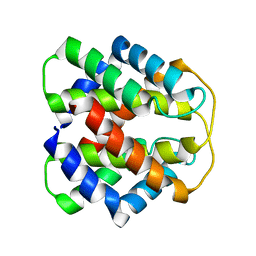 | | Structural Characterization of Minor Ampullate Spidroin Domains and their Distinct Roles in Fibroin Solubility and Fiber Formation | | Descriptor: | Minor ampullate fibroin 1 | | Authors: | Yang, D, Gao, Z, Lin, Z, Huang, W, Lai, C, Fan, J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of minor ampullate spidroin domains and their distinct roles in fibroin solubility and fiber formation
Plos One, 8, 2013
|
|
6AKS
 
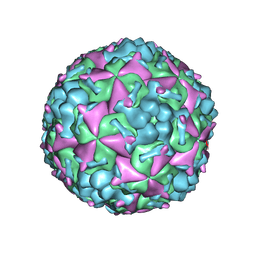 | | Cryo-EM structure of CVA10 mature virus | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKT
 
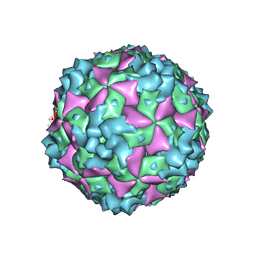 | | Cryo-EM structure of CVA10 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKU
 
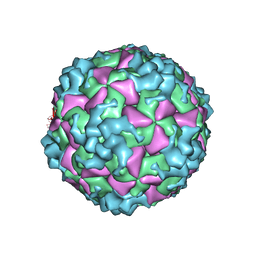 | | Cryo-EM structure of CVA10 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6XK9
 
 | | Cereblon in complex with DDB1, CC-90009, and GSPT1 | | Descriptor: | 2-(4-chlorophenyl)-N-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)-2,2-difluoroacetamide, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | Authors: | Clayton, T.L, Tran, E.T, Zhu, J, Pagarigan, B.E, Matyskiela, M.E, Chamberlain, P.P. | | Deposit date: | 2020-06-25 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells.
Blood, 137, 2021
|
|
4WRQ
 
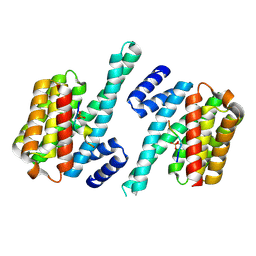 | |
7JVN
 
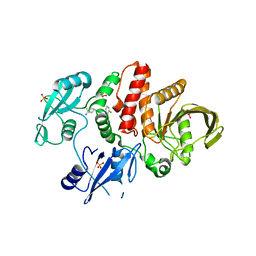 | |
7JVM
 
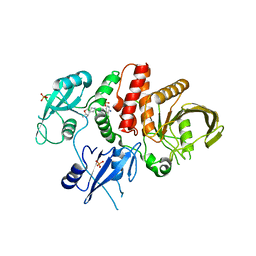 | |
7YOC
 
 | | Crystal structure of Fhb7 | | Descriptor: | Fhb7 | | Authors: | Yang, J, Liang, K, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2022-08-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
3DBC
 
 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 257 | | Descriptor: | 3-[3-(3-methyl-6-{[(1S)-1-phenylethyl]amino}-1H-pyrazolo[4,3-c]pyridin-1-yl)phenyl]propanamide, Polo-like kinase 1 | | Authors: | Elling, R.A, Zhu, J, Barr, K.J, Romanowski, M.J. | | Deposit date: | 2008-05-31 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Design and synthesis of 2-amino-pyrazolopyridines as Polo-like kinase 1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DBF
 
 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 562 | | Descriptor: | 4-({1-[3-(3-amino-3-oxopropyl)-5-chlorophenyl]-3-methyl-1H-pyrazolo[4,3-c]pyridin-6-yl}amino)-3-methoxy-N-(1-methylpipe ridin-4-yl)benzamide, Polo-like kinase | | Authors: | Elling, R.A, Zhu, J, Barr, K.J, Romanowski, M.J. | | Deposit date: | 2008-05-31 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 2-amino-pyrazolopyridines as Polo-like kinase 1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
