2QBY
 
 | | Crystal structure of a heterodimer of Cdc6/Orc1 initiators bound to origin DNA (from S. solfataricus) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 6 homolog 1, Cell division control protein 6 homolog 3, ... | | Authors: | Cunningham Dueber, E.L, Corn, J.E, Bell, S.D, Berger, J.M. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex.
Science, 317, 2007
|
|
3E2L
 
 | | Crystal Structure of the KPC-2 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) | | Descriptor: | Beta-lactamase inhibitory protein, Carbapenemase | | Authors: | Hanes, M.S, Jude, K.M, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2008-08-05 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and biochemical characterization of the interaction between KPC-2 beta-lactamase and beta-lactamase inhibitor protein
Biochemistry, 48, 2009
|
|
3E2K
 
 | | Crystal Structure of the KPC-2 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) | | Descriptor: | Beta-lactamase inhibitory protein, Carbapenemase | | Authors: | Hanes, M.S, Jude, K.M, Berger, J.M, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2008-08-05 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical characterization of the interaction between KPC-2 beta-lactamase and beta-lactamase inhibitor protein
Biochemistry, 48, 2009
|
|
4NMN
 
 | | Aquifex aeolicus replicative helicase (DnaB) complexed with ADP, at 3.3 resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Replicative DNA helicase, ... | | Authors: | Lyubimov, A.Y, Strycharska, M.S, Erzberger, J.P, Berger, J.M. | | Deposit date: | 2013-11-15 | | Release date: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Nucleotide and partner-protein control of bacterial replicative helicase structure and function.
Mol.Cell, 52, 2013
|
|
4GFH
 
 | | Topoisomerase II-DNA-AMPPNP complex | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*CP*GP*TP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*TP*AP*GP*CP*AP*GP*TP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*AP*CP*TP*GP*CP*TP*AP*C)-3'), ... | | Authors: | Schmidt, B.H, Osheroff, N, Berger, J.M. | | Deposit date: | 2012-08-03 | | Release date: | 2012-10-03 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (4.408 Å) | | Cite: | Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FM9
 
 | | Human topoisomerase II alpha bound to DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), DNA topoisomerase 2-alpha, ... | | Authors: | Wendorff, T.J, Schmidt, B.H, Heslop, P, Austin, C.A, Berger, J.M. | | Deposit date: | 2012-06-15 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Structure of DNA-Bound Human Topoisomerase II Alpha: Conformational Mechanisms for Coordinating Inter-Subunit Interactions with DNA Cleavage.
J.Mol.Biol., 424, 2012
|
|
4MN4
 
 | | Structural Basis for the MukB-topoisomerase IV Interaction | | Descriptor: | Chromosome partition protein MukB, DNA topoisomerase 4 subunit A | | Authors: | Vos, S.M, Stewart, N.K, Oakley, M.G, Berger, J.M. | | Deposit date: | 2013-09-09 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the MukB-topoisomerase IV interaction and its functional implications in vivo.
Embo J., 32, 2013
|
|
4KM5
 
 | | X-ray crystal structure of human cyclic GMP-AMP synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Berger, J.M, Doudna, J.A. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity.
Cell Rep, 3, 2013
|
|
3NUH
 
 | |
3NO0
 
 | |
3PEU
 
 | | S. cerevisiae Dbp5 L327V C-terminal domain bound to Gle1 H337R and IP6 | | Descriptor: | ATP-dependent RNA helicase DBP5, GLYCEROL, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3PEW
 
 | | S. cerevisiae Dbp5 L327V bound to RNA and ADP BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3PEV
 
 | | S. cerevisiae Dbp5 L327V C-terminal domain bound to Gle1 and IP6 | | Descriptor: | ATP-dependent RNA helicase DBP5, GLYCEROL, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3PEY
 
 | | S. cerevisiae Dbp5 bound to RNA and ADP BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3RRM
 
 | | S. cerevisiae dbp5 l327v bound to nup159, gle1 h337r, ip6 and adp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Nature, 472, 2011
|
|
3RRN
 
 | | S. cerevisiae dbp5 l327v bound to gle1 h337r and ip6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Nature, 472, 2011
|
|
3R8F
 
 | | Replication initiator DnaA bound to AMPPCP and single-stranded DNA | | Descriptor: | 5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', Chromosomal replication initiator protein dnaA, MAGNESIUM ION, ... | | Authors: | Duderstadt, K.E, Chuang, K, Berger, J.M. | | Deposit date: | 2011-03-23 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.366 Å) | | Cite: | DNA stretching by bacterial initiators promotes replication origin opening.
Nature, 478, 2011
|
|
3TO1
 
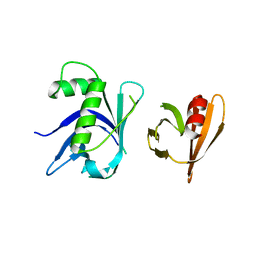 | | Two surfaces on Rtt106 mediate histone binding and chaperone activity | | Descriptor: | Histone chaperone RTT106 | | Authors: | Zunder, R.M, Antczak, A.J, Berger, J.M, Rine, J. | | Deposit date: | 2011-09-02 | | Release date: | 2011-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5JYC
 
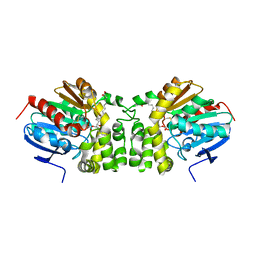 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 14,15-EET hydrolysis intermediate | | Descriptor: | (5~{Z},11~{Z},14~{R},15~{R})-14,15-bis(oxidanyl)icosa-5,8,11-trienoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-05-13 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pseudomonas aeruginosa sabotages the generation of host proresolving lipid mediators.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1EV1
 
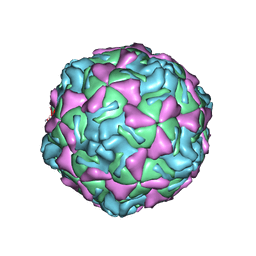 | | ECHOVIRUS 1 | | Descriptor: | ECHOVIRUS 1, MYRISTIC ACID, PALMITIC ACID | | Authors: | Wien, M.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-12-02 | | Release date: | 1999-01-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure determination of echovirus 1.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1JEW
 
 | |
7N85
 
 | | Inner ring spoke from the isolated yeast NPC | | Descriptor: | Nucleoporin ASM4, Nucleoporin NIC96, Nucleoporin NSP1, ... | | Authors: | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | Deposit date: | 2021-06-13 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N84
 
 | | Double nuclear outer ring from the isolated yeast NPC | | Descriptor: | Nucleoporin 145c, Nucleoporin NUP120, Nucleoporin NUP133, ... | | Authors: | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | Deposit date: | 2021-06-13 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | Descriptor: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | Authors: | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
1ESL
 
 | |
