3SW5
 
 | |
3L56
 
 | | Crystal structure of the large c-terminal domain of polymerase basic protein 2 from influenza virus a/viet nam/1203/2004 (h5n1) | | Descriptor: | Polymerase PB2 | | Authors: | Staker, B.L, Edwards, T, Eric, S, Raymond, A, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-12-21 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biological and structural characterization of a host-adapting amino acid in influenza virus.
Plos Pathog., 6, 2010
|
|
8ERR
 
 | |
8ERQ
 
 | |
3EZO
 
 | |
9D32
 
 | |
9DAK
 
 | |
9E0I
 
 | |
7JX3
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab domain of monoclonal antibody S2H14, Heavy chain of Fab domain of monoclonal antibody S304, ... | | Authors: | Snell, G, Czudnochowski, N, Rosen, L.E, Nix, J.C, Corti, D, Veesler, D, Park, Y.J, Walls, A.C, Tortorici, M.A, Cameroni, E, Pinto, D, Beltramello, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-08-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
9EH8
 
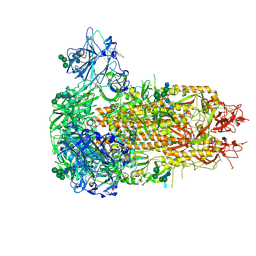 | | Structure of the prefusion HKU5-19s Spike trimer (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Gen, R, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-11-22 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Molecular basis of convergent evolution of ACE2 receptor utilization among HKU5 coronaviruses.
Cell, 188, 2025
|
|
9EA0
 
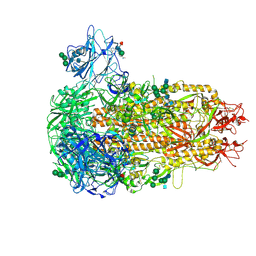 | | Structure of the prefusion HKU5-19s Spike trimer (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Gen, R, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-11-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Molecular basis of convergent evolution of ACE2 receptor utilization among HKU5 coronaviruses.
Cell, 188, 2025
|
|
3TCQ
 
 | | Crystal Structure of matrix protein VP40 from Ebola virus Sudan | | Descriptor: | Matrix protein VP40 | | Authors: | Clifton, M.C, Edwards, T.E, Anderson, V, Atkins, K, Raymond, A, Saphire, E.O, Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution Crystal Structure of Dimeric VP40 From Sudan ebolavirus.
J Infect Dis, 212 Suppl 2, 2015
|
|
7M53
 
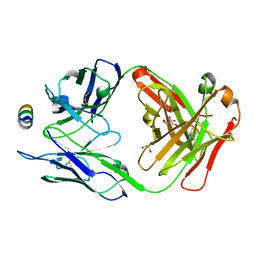 | | B6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M55
 
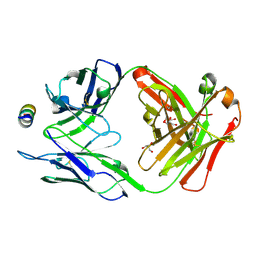 | | B6 Fab fragment bound to the MERS-CoV spike stem helix peptide | | Descriptor: | B6 antigen binding fragment (Fab) heavy chain, B6 antigen binding fragment (Fab) light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M52
 
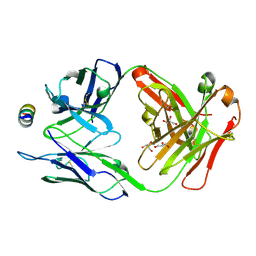 | | B6 Fab fragment bound to the HKU4 spike stem helix peptide | | Descriptor: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3T1D
 
 | | The mutant structure of human Siderocalin W79A, R81A, Y106F bound to Enterobactin | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-07-21 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Parsing the functional specificity of Siderocalin/Lipocalin 2/NGAL for siderophores and related small-molecule ligands.
J Struct Biol X, 2, 2019
|
|
2MYY
 
 | |
7TN0
 
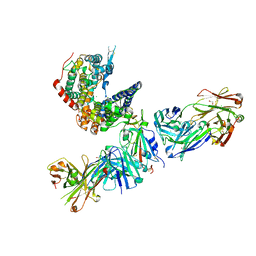 | | SARS-CoV-2 Omicron RBD in complex with human ACE2 and S304 Fab and S309 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | McCallum, M, Czudnochowski, N, Nix, J.C, Croll, T.I, SSGCID, Dillen, J.R, Snell, G, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement.
Science, 375, 2022
|
|
9DNE
 
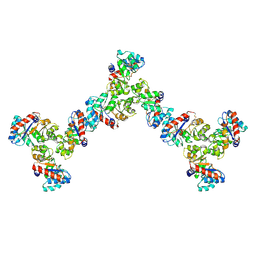 | | Pseudosymmetric protein nanocage GI9-F7 (local refinement) | | Descriptor: | Pseudosymmetric protein nanocage GI9-F7 A chain, Pseudosymmetric protein nanocage GI9-F7 B chain, Pseudosymmetric protein nanocage GI9-F7 C chain | | Authors: | Park, Y.J, Dowling, Q.M, King, N.P, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-09-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Hierarchical design of pseudosymmetric protein nanocages.
Nature, 638, 2025
|
|
9DND
 
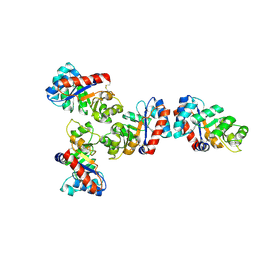 | | Pseudosymmetric protein nanocage GI4 -F7 (local refinement) | | Descriptor: | Pseudosymmetric protein nanocages GI4 A Chain, Pseudosymmetric protein nanocages GI4 B Chain, Pseudosymmetric protein nanocages GI4 C Chain | | Authors: | Park, Y.J, Dowling, Q.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID), King, N.P, Veesler, D. | | Deposit date: | 2024-09-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Hierarchical design of pseudosymmetric protein nanocages.
Nature, 638, 2025
|
|
9C6O
 
 | |
8U29
 
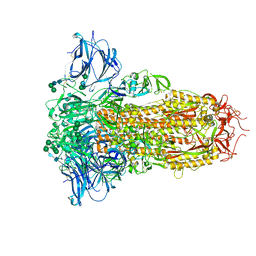 | | Prefusion structure of the PRD-0038 spike glycoprotein ectodomain trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PRD-0038 Spike glycoprotein, ... | | Authors: | Lee, J, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-09-05 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Broad receptor tropism and immunogenicity of a clade 3 sarbecovirus.
Cell Host Microbe, 31, 2023
|
|
9DGO
 
 | |
9DB3
 
 | |
9DB1
 
 | |
