3FK9
 
 | | Crystal structure of mMutator MutT protein from Bacillus halodurans | | Descriptor: | Mutator MutT protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of mMutator MutT protein from Bacillus halodurans
To be Published
|
|
3EWD
 
 | | Crystal structure of adenosine deaminase mutant (delta Asp172) from Plasmodium vivax in complex with MT-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, Adenosine deaminase, ZINC ION | | Authors: | Schramm, V.L, Almo, S.C, Cassera, M.B, Ho, M.C. | | Deposit date: | 2008-10-14 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and metabolic specificity of methylthiocoformycin for malarial adenosine deaminases.
Biochemistry, 48, 2009
|
|
3FBT
 
 | | Crystal structure of a chorismate mutase/shikimate 5-dehydrogenase fusion protein from Clostridium acetobutylicum | | Descriptor: | SULFATE ION, chorismate mutase and shikimate 5-dehydrogenase fusion protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a chorismate mutase/shikimate 5-dehydrogenase fusion protein from Clostridium acetobutylicum
To be Published
|
|
3EVY
 
 | | Crystal structure of a fragment of a putative type I restriction enzyme R protein from Bacteroides fragilis | | Descriptor: | Putative type I restriction enzyme R protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a fragment of a putative type I restriction enzyme R protein from Bacteroides fragilis
To be Published
|
|
3F13
 
 | | Crystal structure of putative nudix hydrolase family member from Chromobacterium violaceum | | Descriptor: | putative nudix hydrolase family member | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative nudix hydrolase family member from Chromobacterium violaceum
To be Published
|
|
3FEF
 
 | | Crystal structure of putative glucosidase lplD from bacillus subtilis | | Descriptor: | MAGNESIUM ION, Putative glucosidase lplD, ALPHA-GALACTURONIDASE, ... | | Authors: | Ramagopal, U.A, Rajashankar, K.R, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative glucosidase lplD from bacillus subtilis.
To be published
|
|
3FOM
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide IQQSIERL | | Descriptor: | 8 residue synthetic peptide, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Brims, D.R, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Malashkevich, V.N, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect.
Int.Immunol., 22, 2010
|
|
3FK4
 
 | | Crystal structure of RuBisCO-like protein from Bacillus Cereus ATCC 14579 | | Descriptor: | RuBisCO-like protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-15 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of RuBisCO-like protein from Bacillus Cereus ATCC 14579
To be Published
|
|
3EVT
 
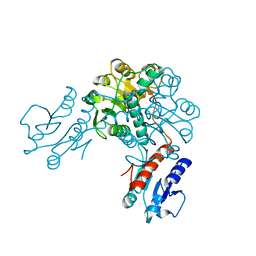 | | Crystal structure of phosphoglycerate dehydrogenase from Lactobacillus plantarum | | Descriptor: | Phosphoglycerate dehydrogenase | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phosphoglycerate dehydrogenase from Lactobacillus plantarum
To be Published
|
|
3F5Q
 
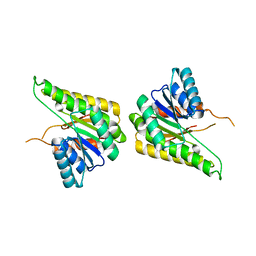 | | CRYSTAL STRUCTURE OF putative short chain dehydrogenase from Escherichia coli CFT073 | | Descriptor: | dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of an uncharacterized protein
to be published
|
|
3CG0
 
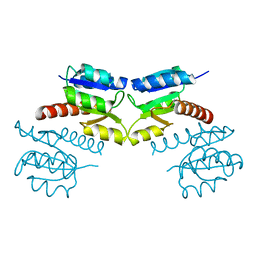 | | Crystal structure of signal receiver domain of modulated diguanylate cyclase from Desulfovibrio desulfuricans G20, an example of alternate folding | | Descriptor: | Response regulator receiver modulated diguanylate cyclase with PAS/PAC sensor | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Chang, S, Groshong, C, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Modulated Diguanylate Cyclase from Desulfovibrio desulfuricans.
To be Published
|
|
3DGR
 
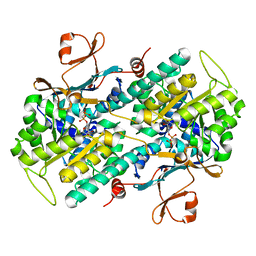 | | Crystal structure of human NAMPT complexed with ADP analogue | | Descriptor: | Nicotinamide phosphoribosyltransferase, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Ho, M, Burgos, E.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2008-06-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3DHF
 
 | | Crystal structure of phosphorylated mimic form of human NAMPT complexed with nicotinamide mononucleotide and pyrophosphate | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ho, M, Burgos, E.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2008-06-17 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3DUT
 
 | | The high salt (phosphate) crystal structure of deoxy hemoglobin E (GLU26LYS) at physiological pH (pH 7.35) | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PHOSPHATE ION, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2008-07-17 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The high salt (phosphate) crystal structure of deoxy
hemoglobin E (GLU26LYS) at physiological pH (pH 7.35)
To be Published
|
|
3DO9
 
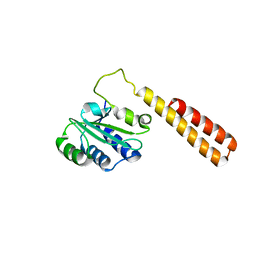 | | Crystal structure of protein ba1542 from bacillus anthracis str.ames | | Descriptor: | UPF0302 protein BA_1542/GBAA1542/BAS1430 | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Iizuka, M, Maletic, M, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-03 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Protein Ba1542 from Bacillus Anthracis Str.Ames.
To be Published
|
|
3DUG
 
 | | Crystal structure of zn-dependent arginine carboxypeptidase complexed with zinc | | Descriptor: | ARGININE, GLYCEROL, ZINC ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Freeman, J, Iizuka, M, Bain, K, Rodgers, L, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-17 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes.
Biochemistry, 48, 2009
|
|
3D4P
 
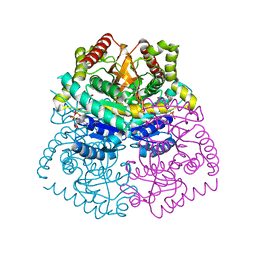 | |
3DBI
 
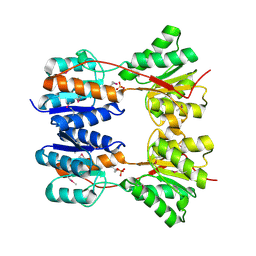 | | CRYSTAL STRUCTURE OF SUGAR-BINDING TRANSCRIPTIONAL REGULATOR (LACI FAMILY) FROM ESCHERICHIA COLI COMPLEXED WITH PHOSPHATE | | Descriptor: | GLYCEROL, PHOSPHATE ION, SUGAR-BINDING TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Wu, B, Maletic, M, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-01 | | Release date: | 2008-07-01 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Sugar-Binding Transcriptional Regulator (LacI Family) from Escherichia Coli Complexed with Phosphate.
To be Published
|
|
3DES
 
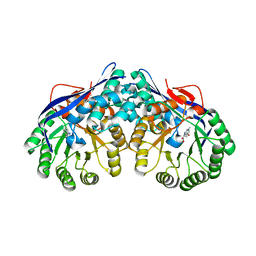 | | Crystal structure of dipeptide epimerase from Thermotoga maritima complexed with L-Ala-L-Phe dipeptide | | Descriptor: | ALANINE, MAGNESIUM ION, Muconate cycloisomerase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening.
Structure, 16, 2008
|
|
3DG6
 
 | | Crystal structure of muconate lactonizing enzyme from Mucobacterium Smegmatis complexed with muconolactone | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, [(2S)-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes
Biochemistry, 48, 2009
|
|
3DBY
 
 | | Crystal structure of uncharacterized protein from Bacillus cereus G9241 (CSAP Target) | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, uncharacterized protein | | Authors: | Ramagopal, U.A, Bonanno, J.B, Ozyurt, S, Freeman, J, Wasserman, S, Hu, S, Groshong, C, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-29 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of uncharacterized protein from Bacillus cereus G9241
To be published
|
|
3DG7
 
 | | Crystal structure of muconate lactonizing enzyme from Mucobacterium Smegmatis complexed with muconolactone | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, [(2S)-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes
Biochemistry, 48, 2009
|
|
3DER
 
 | | Crystal structure of dipeptide epimerase from Thermotoga maritima complexed with L-Ala-L-Lys dipeptide | | Descriptor: | ALANINE, LYSINE, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening.
Structure, 16, 2008
|
|
3FRN
 
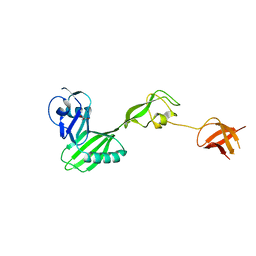 | | CRYSTAL STRUCTURE OF flagellar protein FlgA FROM Thermotoga maritima MSB8 | | Descriptor: | Flagellar protein FlgA, GLYCEROL | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Hu, S, Bain, K, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CRYSTAL STRUCTURE OF flagellar protein FlgA FROM Thermotoga maritima
To be Published
|
|
3G17
 
 | |
