6XMK
 
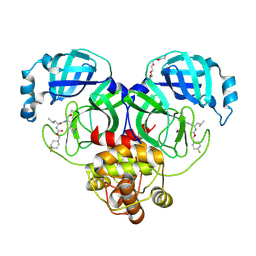 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
2RJZ
 
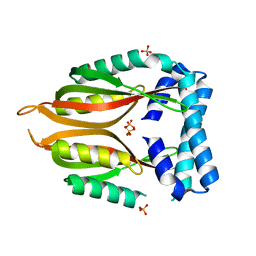 | | Crystal structure of the type 4 fimbrial biogenesis protein PilO from Pseudomonas aeruginosa | | Descriptor: | PilO protein, SULFATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-16 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Periplasmic domains of Pseudomonas aeruginosa PilN and PilO form a stable heterodimeric complex.
J.Mol.Biol., 394, 2009
|
|
8I2Z
 
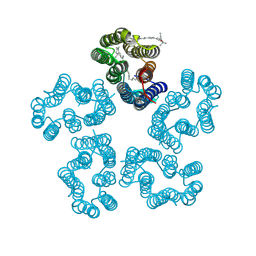 | | Cryo-EM structure of the zeaxanthin-bound kin4B8 | | Descriptor: | RETINAL, Xanthorhodopsin, Zeaxanthin | | Authors: | Murakoshi, S, Chazan, A, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2023-01-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Phototrophy by antenna-containing rhodopsin pumps in aquatic environments.
Nature, 615, 2023
|
|
5WB6
 
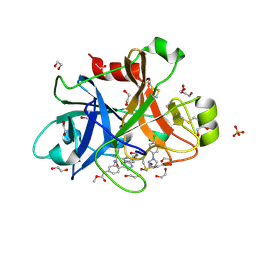 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(11S)-11-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-6-fluoro-2-oxo-1,3,4,10,11,13-hexahydro-2H-5,9:15,12-di(azeno)-1,13-benzodiazacycloheptadecin-18-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrocyclic factor XIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6DXL
 
 | | Linked amidobenzimidazole STING agonist | | Descriptor: | 1,1'-(butane-1,4-diyl)bis{2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1H-benzimidazole-5-carboxamide}, CALCIUM ION, Stimulator of interferon protein | | Authors: | Concha, N.O. | | Deposit date: | 2018-06-29 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
6DXG
 
 | | amidobenzimidazole (ABZI) STING agonists | | Descriptor: | 2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1-[(2R)-2-hydroxy-2-phenylethyl]-1H-benzimidazole-5-carboxamide, CALCIUM ION, Stimulator of interferon protein | | Authors: | Concha, N.O. | | Deposit date: | 2018-06-28 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
4USD
 
 | | Human STK10 (LOK) with SB-633825 | | Descriptor: | 4-{5-(6-methoxynaphthalen-2-yl)-1-methyl-2-[2-methyl-4-(methylsulfonyl)phenyl]-1H-imidazol-4-yl}pyridine, SERINE/THREONINE-PROTEIN KINASE 10 | | Authors: | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Canning, P, Raynor, J, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
4USF
 
 | | Human SLK with SB-440719 | | Descriptor: | 4-[4-(6-methoxynaphthalen-2-yl)-1H-imidazol-5-yl]pyridine, STE20-LIKE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
7N2B
 
 | | A DARPin semi-rigidly fused to the 3TEL crystallization chaperone | | Descriptor: | Transcription factor ETV6,Transcription factor ETV6,Transcription factor ETV6,3TEL-rigid-DARPin | | Authors: | Sarath Nawarathange, S.D, Gajjar, P, Bunn, D, Stewart, C, Doukov, T, Moody, J.D. | | Deposit date: | 2021-05-28 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.221 Å) | | Cite: | Crystals of TELSAM-target protein fusions that exhibit minimal crystal contacts and lack direct inter-TELSAM contacts.
Open Biology, 12, 2022
|
|
7N1O
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Transcription factor ETV6,Isoform 4 of Anthrax toxin receptor 2 | | Authors: | Mathis, M.H, Bezzant, B.D, Ramirez, D.T, Sarath Nawarathnage, S.D, Doukov, T, Moody, J.D. | | Deposit date: | 2021-05-27 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystals of TELSAM-target protein fusions that exhibit minimal crystal contacts and lack direct inter-TELSAM contacts.
Open Biology, 12, 2022
|
|
4ZLO
 
 | | Serine/threonine-protein kinase PAK1 complexed with a dibenzodiazepine: identification of an allosteric site on PAK1 | | Descriptor: | 2,8-difluoro-11-(4-methylpiperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine, GLYCEROL, Serine/threonine-protein kinase PAK 1 | | Authors: | Bellamacina, C.R, Bussiere, D.E. | | Deposit date: | 2015-05-01 | | Release date: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization of a Dibenzodiazepine Hit to a Potent and Selective Allosteric PAK1 Inhibitor.
Acs Med.Chem.Lett., 6, 2015
|
|
5WKL
 
 | | 1.85 A resolution structure of MERS 3CL protease in complex with piperidine-based peptidomimetic inhibitor 17 | | Descriptor: | (1R,2S)-2-{[N-({[4-benzyl-1-(tert-butoxycarbonyl)piperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[4-benzyl-1-(tert-butoxycarbonyl)piperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
2GPA
 
 | | ALLOSTERIC INHIBITION OF GLYCOGEN PHOSPHORYLASE A BY A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oikonomakos, N.G, Tsitsanou, K.E, Zographos, S.E, Skamnaki, V.T. | | Deposit date: | 1999-02-18 | | Release date: | 1999-02-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric inhibition of glycogen phosphorylase a by the potential antidiabetic drug 3-isopropyl 4-(2-chlorophenyl)-1,4-dihydro-1-ethyl-2-methyl-pyridine-3,5,6-tricarbo xylate.
Protein Sci., 8, 1999
|
|
4ZPN
 
 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 with extended N-terminus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MCG133388, ... | | Authors: | Goodman, K.M, Wolcott, H.N, Bahna, F, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
5VR1
 
 | |
5W0I
 
 | | CREBBP Bromodomain in complex with Cpd8 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
1MZG
 
 | | X-Ray Structure of SufE from E.coli Northeast Structural Genomics (NESG) Consortium Target ER30 | | Descriptor: | SufE Protein | | Authors: | Kuzin, A, Edstrom, W.C, Xiao, R, Acton, T.B, Rost, B, Tong, L, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-07 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes
J.Mol.Biol., 344, 2004
|
|
3LOX
 
 | | HCV NS3-4a protease domain with a ketoamide inhibitor derivative of Boceprevir bound | | Descriptor: | (1R,2S,5S)-N-[(2S,3R)-4-amino-1-cyclobutyl-3-hydroxy-4-oxobutan-2-yl]-6,6-dimethyl-3-{3-methyl-N-[(1-methylcyclohexyl)c arbamoyl]-L-valyl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, BETA-MERCAPTOETHANOL, HCV NS3 Protease, ... | | Authors: | Prongay, A.J. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The introduction of P4 substituted 1-methylcyclohexyl groups into Boceprevir: a change in direction in the search for a second generation HCV NS3 protease inhibitor.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5I89
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02857790 | | Descriptor: | (4R)-6-(3-cyclopropyl-1-methyl-1H-indazol-5-yl)-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Setser, J.W, Poy, F, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
7D40
 
 | |
5I83
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02773986 | | Descriptor: | (4R)-4-methyl-7-[(1R)-1-phenylethoxy]-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, CREB-binding protein, THIOCYANATE ION | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
7D96
 
 | |
7D97
 
 | |
7D95
 
 | |
5IML
 
 | | Nanobody targeting human Vsig4 in Spacegroup P212121 | | Descriptor: | Nanobody, V-set and immunoglobulin domain-containing protein 4 | | Authors: | Wen, Y. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
