4EH1
 
 | | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoprotein, ... | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
4E4R
 
 | | EutD phosphotransacetylase from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | EutD phosphotransacetylase from Staphylococcus aureus.
To be Published
|
|
1NEZ
 
 | | The Crystal Structure of a TL/CD8aa Complex at 2.1A resolution:Implications for Memory T cell Generation, Co-receptor Preference and Affinity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, Y, Xiong, Y, Naidenko, O.V, Liu, J.H, Zhang, R, Joachimiak, A, Kronenberg, M, Cheroutre, H, Reinherz, E.L, Wang, J.H. | | Deposit date: | 2002-12-12 | | Release date: | 2003-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a TL/CD8alphaalpha Complex at 2.1 A resolution: Implications for modulation of T cell activation and memory
Immunity, 18, 2003
|
|
1NC5
 
 | | Structure of Protein of Unknown Function of YteR from Bacillus Subtilis | | Descriptor: | hypothetical protein yTER | | Authors: | Zhang, R, Lozondra, L, Korolev, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6 A crystal structure of YteR protein from Bacillus subtilis, a predicted lyase.
Proteins, 60, 2005
|
|
1NG6
 
 | | Structure of Cytosolic Protein of Unknown Function YqeY from Bacillus subtilis | | Descriptor: | Hypothetical protein yqeY | | Authors: | Zhang, R, Dementiva, I, Vinokour, E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-16 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4A crystal structure of hypothetical cytosolic protein
YQEY
To be Published
|
|
4EJO
 
 | | Crystal structure of padr family transcriptional regulator from Eggerthella lenta DSM 2243 | | Descriptor: | ETHANOL, Transcriptional regulator, PadR-like family | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of padr family transcriptional regulator from Eggerthella lenta DSM 2243
TO BE PUBLISHED
|
|
4GS5
 
 | | The crystal structure of acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein from Dyadobacter fermentans DSM 18053 | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein, IODIDE ION | | Authors: | Tan, K, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-27 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | The crystal structure of acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein from Dyadobacter fermentans DSM 18053
To be Published
|
|
4GXT
 
 | | The crystal structure of a conserved functionally unknown protein from Anaerococcus prevotii DSM 20548 | | Descriptor: | GLYCEROL, SULFATE ION, a conserved functionally unknown protein | | Authors: | Tan, K, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | The crystal structure of a conserved functionally unknown protein from Anaerococcus prevotii DSM 20548
To be Published
|
|
3U7I
 
 | | The crystal structure of FMN-dependent NADH-azoreductase 1 (GBAA0966) from Bacillus anthracis str. Ames Ancestor | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Zhang, R, Gu, M, Tan, K, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-13 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of FMN-dependent NADH-azoreductase 1 (GBAA0966) from Bacillus anthracis str. Ames Ancestor
To be Published
|
|
3TY6
 
 | | ATP-dependent Protease HslV from Bacillus anthracis str. Ames | | Descriptor: | ATP-dependent protease subunit HslV, SULFATE ION | | Authors: | Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-23 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | ATP-dependent Protease HslV from Bacillus anthracis str. Ames
To be Published
|
|
3TV9
 
 | | Crystal Structure of Putative Peptide Maturation Protein from Shigella flexneri | | Descriptor: | GLYCEROL, Putative peptide maturation protein, SULFATE ION | | Authors: | Kim, Y, Maltseva, N, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-19 | | Release date: | 2011-10-05 | | Last modified: | 2016-12-28 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal Structure of Putative Peptide Maturation Protein from
To be Published
|
|
4HN3
 
 | | The crystal structure of a sex pheromone precursor (lmo1757) from Listeria monocytogenes EGD-e | | Descriptor: | BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-18 | | Release date: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | The crystal structure of a sex pheromone precursor (lmo1757) from Listeria monocytogenes EGD-e
To be Published
|
|
5UOU
 
 | | High resolution structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) decarboxylase | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-01 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To Be Published
|
|
5UPS
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis in complex with SBNP663 ligand | | Descriptor: | 5'-O-[(R)-hydroxy{[(7beta,8alpha,9beta,10alpha,11beta,13alpha)-7-hydroxy-19-oxo-11,16-epoxykauran-19-yl]oxy}phosphoryl]adenosine, Acyl-CoA synthetase PtmA2, FORMIC ACID, ... | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Chang, C.-Y, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
3USB
 
 | | Crystal Structure of Bacillus anthracis Inosine Monophosphate Dehydrogenase in the complex with IMP | | Descriptor: | CHLORIDE ION, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Zhang, R, Wu, R, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
5ULB
 
 | |
4F3U
 
 | |
4F66
 
 | | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutans UA159 in complex with beta-D-glucose-6-phosphate. | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Tan, K, Michalska, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4F4I
 
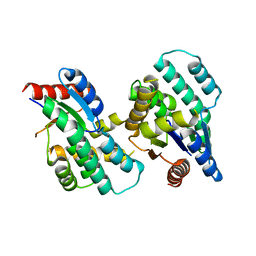 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in apo-form | | Descriptor: | Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-05-10 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in apo-form
To be Published
|
|
4F79
 
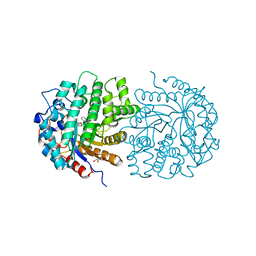 | | The crystal structure of 6-phospho-beta-glucosidase mutant (E375Q) in complex with Salicin 6-phosphate | | Descriptor: | 2-(hydroxymethyl)phenyl 6-O-phosphono-beta-D-glucopyranoside, GLYCEROL, Putative phospho-beta-glucosidase | | Authors: | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
3UKJ
 
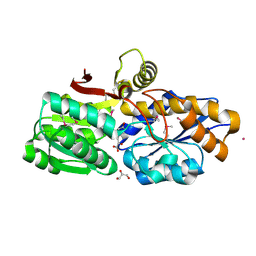 | | Crystal structure of extracellular ligand-binding receptor from Rhodopseudomonas palustris HaA2 | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, GLYCEROL, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-09 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4EYQ
 
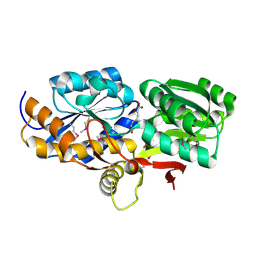 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 in complex with caffeic acid/3-(4-HYDROXY-PHENYL)PYRUVIC ACID | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, CAFFEIC ACID, Extracellular ligand-binding receptor | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4I4K
 
 | | Streptomyces globisporus C-1027 9-membered enediyne conserved protein SgcE6 | | Descriptor: | CITRIC ACID, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J Antibiot (Tokyo), 69, 2016
|
|
4HKY
 
 | | New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Faropenem | | Descriptor: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, (5R,6S)-6-(1-hydroxyethyl)-7-oxo-3-[(2R)-oxolan-2-yl]-4-thia-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-10-15 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Faropenem
To be Published
|
|
4HX6
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 | | Descriptor: | ACETATE ION, Oxidoreductase, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-09 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
