6CNG
 
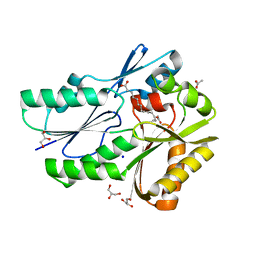 | | The X-ray crystal structure of the Streptococcus pneumoniae Fatty Acid Kinase (Fak) B3 protein loaded with linoleic acid to 1.47 Angstrom resolution | | Descriptor: | ACETATE ION, Fatty Acid Kinase (Fak) B3 protein, GLYCEROL, ... | | Authors: | Cuypers, M.G, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2018-03-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The X-ray crystal structure of the Streptococcus pneumoniae Fatty Acid Kinase (Fak) B3 protein loaded with linoleic acid to 1.47 Angstrom resolution
To Be Published
|
|
2FYW
 
 | |
8PCZ
 
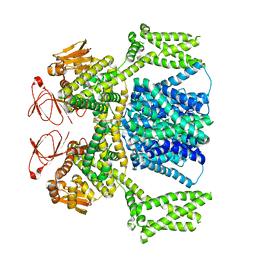 | | Ligand-free SpSLC9C1 in lipid nanodiscs, dimer | | Descriptor: | Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PD2
 
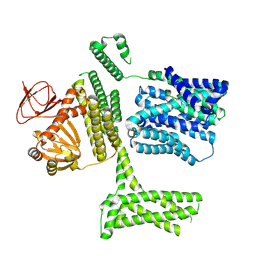 | | Ligand-free SpSLC9C1 in lipid nanodiscs, protomer state 1 | | Descriptor: | Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-11 | | Release date: | 2023-11-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
2G6G
 
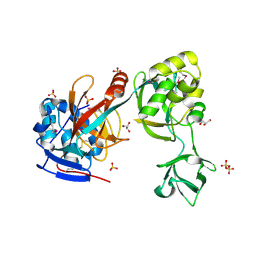 | | Crystal structure of MltA from Neisseria gonorrhoeae | | Descriptor: | GLYCEROL, GNA33, SULFATE ION | | Authors: | Powell, A.J, Liu, Z.J, Nicholas, R.A, Davies, C. | | Deposit date: | 2006-02-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of the Lytic Transglycosylase MltA from N.gonorrhoeae and E.coli: Insights into Interdomain Movements and Substrate Binding.
J.Mol.Biol., 359, 2006
|
|
8PD7
 
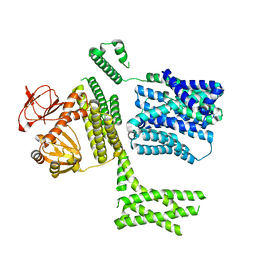 | | Ligand-free SpSLC9C1 in lipid nanodiscs, protomer state 4 | | Descriptor: | Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
6UYV
 
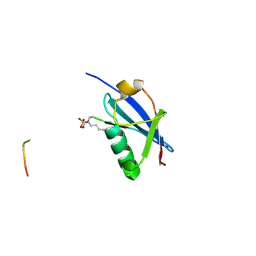 | | Crystal structure of K46-acetylated SUMO1 in complex with phosphorylated PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
4UHH
 
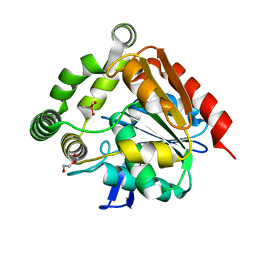 | | Structural studies of a thermophilic esterase from Thermogutta terrifontis (cacodylate complex) | | Descriptor: | CACODYLIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Isupov, M.N, Bonch-Osmolovskaya, E, Littlechild, J.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural Studies of a Thermophilic Esterase from a New Planctomycetes Species, Thermogutta Terrifontis.
FEBS J., 282, 2015
|
|
8PD5
 
 | | Ligand-free SpSLC9C1 in lipid nanodiscs, protomer state 3 | | Descriptor: | Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
2G01
 
 | | Pyrazoloquinolones as Novel, Selective JNK1 inhibitors | | Descriptor: | 6-CHLORO-9-HYDROXY-1,3-DIMETHYL-1,9-DIHYDRO-4H-PYRAZOLO[3,4-B]QUINOLIN-4-ONE, C-jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-02-10 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Synthesis and SAR of 1,9-dihydro-9-hydroxypyrazolo[3,4-b]quinolin-4-ones as novel, selective c-Jun N-terminal kinase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8PDU
 
 | | cGMP-bound SpSLC9C1 in lipid nanodiscs, dimer | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PD9
 
 | | cAMP-bound SpSLC9C1 in lipid nanodiscs, protomer state 1 | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PDV
 
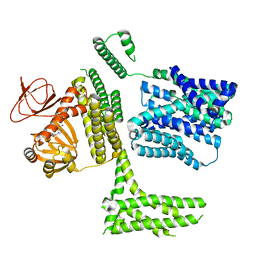 | | cGMP-bound SpSLC9C1 in lipid nanodiscs, protomer | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
6CBE
 
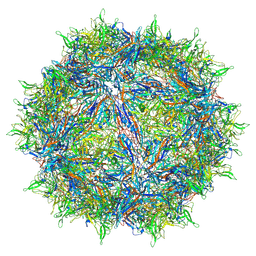 | | Atomic structure of a rationally engineered gene delivery vector, AAV2.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Burg, M, Rosebrough, C, Drouin, L, Bennett, A, Mietzsch, M, Chipman, P, McKenna, R, Sousa, D, Potter, M, Byrne, B, Kozyreva, O.G, Samulski, R.J, Agbandje-McKenna, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-05-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Atomic structure of a rationally engineered gene delivery vector, AAV2.5.
J. Struct. Biol., 203, 2018
|
|
8PC2
 
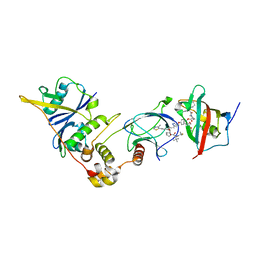 | | SelDeg51 in complex with FKBP51FK1 domain and pVHL:EloB:EloC | | Descriptor: | Elongin-B, Elongin-C, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Meyners, C, Walz, M, Geiger, T.M, Hausch, F. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Potent Proteolysis Targeting Chimera Enables Targeting the Scaffolding Functions of FK506-Binding Protein 51 (FKBP51).
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6CIG
 
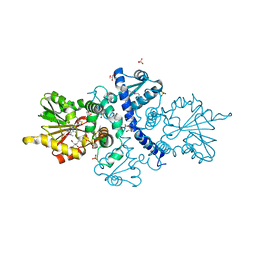 | | CRYSTAL STRUCTURE ANALYSIS OF SELENOMETHIONINE SUBSTITUTED ISOFLAVONE O-METHYLTRANSFERASE | | Descriptor: | GLYCEROL, Isoflavone-7-O-methyltransferase 8, N-(TRIS(HYDROXYMETHYL)METHYL)-3-AMINOPROPANESULFONIC ACID, ... | | Authors: | Zubieta, C, Dixon, R.A, Shabalin, I.G, Kowiel, M, Porebski, P.J, Noel, J.P. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat. Struct. Biol., 8, 2001
|
|
8PDF
 
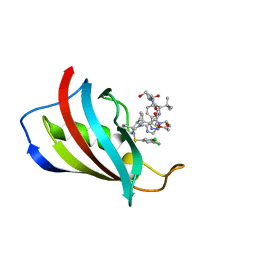 | | FKBP12 in complex with PROTAC 6a2 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[4-[(1~{S})-1-[(1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-2-oxidanylidene-3,10-diazabicyclo[4.3.1]decan-3-yl]ethyl]-1,2,3-triazol-1-yl]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Meyners, C, Walz, M, Geiger, T.M, Hausch, F. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of a Potent Proteolysis Targeting Chimera Enables Targeting the Scaffolding Functions of FK506-Binding Protein 51 (FKBP51).
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
2GDN
 
 | |
8OXA
 
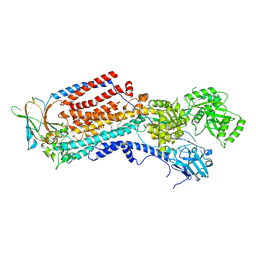 | | Cryo-EM structure of ATP8B1-CDC50A in E2-Pi conformation with occluded PS | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
6V1I
 
 | | Cryo-EM reconstruction of the thermophilic bacteriophage P74-26 small terminase- symmetric | | Descriptor: | Small terminase protein | | Authors: | Hayes, J.A, Hilbert, B.J, Gaubitz, C, Stone, N.P, Kelch, B.A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A thermophilic phage uses a small terminase protein with a fixed helix-turn-helix geometry.
J.Biol.Chem., 295, 2020
|
|
6V1V
 
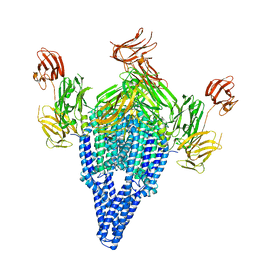 | | VIP3B (VIP3B_2160) adapted for crystallization | | Descriptor: | Vegetative insecticidal protein | | Authors: | Evdokimov, A.G, Zheng, M, Moshiri, F, Haas, J, Lowder, C. | | Deposit date: | 2019-11-21 | | Release date: | 2019-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.189 Å) | | Cite: | Crystal structure of a Vip3B family insecticidal protein reveals a new fold and a unique tetrameric assembly.
Protein Sci., 29, 2020
|
|
8OX7
 
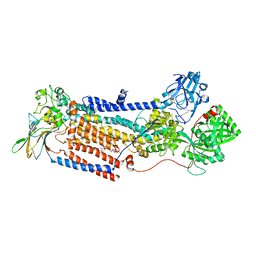 | | Cryo-EM structure of ATP8B1-CDC50A in E2P autoinhibited "closed" conformation | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
6CLA
 
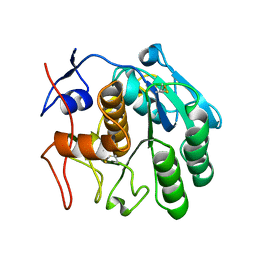 | | 2.80 A MicroED structure of proteinase K at 6.0 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6V6Y
 
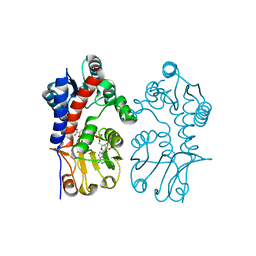 | | Crystal Structure of T. thermophilus methylenetetrahydrofolate dehydrogenase (MTHFD) | | Descriptor: | Bifunctional protein FolD, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Maiello, F, Coelho, C, Gallo, G, Sucharski, F, Hardy, L, Wurtele, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15201569 Å) | | Cite: | Crystal structure of Thermus thermophilus methylenetetrahydrofolate dehydrogenase and determinants of thermostability.
Plos One, 15, 2020
|
|
6CLJ
 
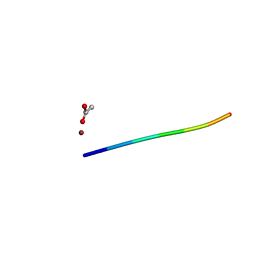 | | 1.01 A MicroED structure of GSNQNNF at 0.50 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
