6A0O
 
 | | Mandelate oxidase mutant-Y128F with benzaldehyde | | Descriptor: | 4-hydroxymandelate oxidase, FLAVIN MONONUCLEOTIDE, benzaldehyde | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical and structural explorations of alpha-hydroxyacid oxidases reveal a four-electron oxidative decarboxylation reaction.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A0Y
 
 | | Mandelate oxidase mutant-Y128F with Benzoic acid | | Descriptor: | 4-hydroxymandelate oxidase, BENZOIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural explorations of alpha-hydroxyacid oxidases reveal a four-electron oxidative decarboxylation reaction.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A1H
 
 | | Mandelate oxidase mutant-Y128F with 5-deazariboflavin mononucleotide | | Descriptor: | 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A0M
 
 | | Mandelate oxidase mutant-Y128F with 2-Phenylacetic acid | | Descriptor: | 2-PHENYLACETIC ACID, 4-hydroxymandelate oxidase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural explorations of alpha-hydroxyacid oxidases reveal a four-electron oxidative decarboxylation reaction.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A1R
 
 | | Mandelate oxidase mutant-Y128F with the N5-phenylacetyl-FMN adduct | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-2,4-dioxo-5-(phenylacetyl)-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase, MAGNESIUM ION | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A1L
 
 | | Mandelate oxidase mutant-Y128F with 5-deazariboflavin mononucleotide and benzoic acid | | Descriptor: | 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase, BENZOIC ACID, ... | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A3T
 
 | | The crystal structure of Mandelate oxidase R163L with 2-hydroxy-phenylacetamide | | Descriptor: | 1-{5-[(1S)-2-amino-1-hydroxy-2-oxo-1-phenylethyl]-7,8-dimethyl-2,4-dioxo-1,2,3,4-tetrahydrobenzo[g]pteridine-5,10-diium-10-yl}-1-deoxy-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-16 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.511 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7EBO
 
 | |
8I80
 
 | | Crystal structure of Cph001-D189N | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J. | | Deposit date: | 2023-02-02 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I86
 
 | | Crystal structure of Cph001-D189N in complex with GTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-TRIPHOSPHATE, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I84
 
 | | Crystal structure of Cph001-D189N in complex with CMN IIB | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, KBE-DPP-UAL-MYN-DPP-ALA, ... | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I85
 
 | | Crystal structure of Cph001-D189N in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I89
 
 | | Crystal structure of Cph001-D189N in complex with VIO | | Descriptor: | DI(HYDROXYETHYL)ETHER, KBE-DPP-SER-SER-UAL-5OH, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I82
 
 | |
8I8G
 
 | | Crystal structure of Cph001-D189N in complex with CMN IIA and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, KBE-DPP-UAL-MYN-DPP-SER, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-04 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I8H
 
 | | Crystal structure of Cph001-D189N in complex with VIO and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, KBE-DPP-SER-SER-UAL-5OH, ... | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-04 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
7VYY
 
 | | The crystal structure of Non-hydrolyzing UDPGlcNAc 2-epimerase | | Descriptor: | Putative UDP-N-acetylglucosamine 2-epimerase, SODIUM ION | | Authors: | Li, T.L, Rattinam, R. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.443 Å) | | Cite: | KasQ an Epimerase Primes the Biosynthesis of Aminoglycoside Antibiotic Kasugamycin and KasF/H Acetyltransferases Inactivate Its Activity.
Biomedicines, 10, 2022
|
|
7VZA
 
 | |
7VZ6
 
 | |
6IIY
 
 | |
6IIX
 
 | |
5YDG
 
 | | Crystal structure of the Arabidopsis thaliana chloroplast RNA editing factors 2(MORF2) | | Descriptor: | Multiple organellar RNA editing factor 2, chloroplastic | | Authors: | Wang, X, Yang, J.Y, Wang, Y.L, Gao, Y.S. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal structure of the chloroplast RNA editing factor MORF2
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5Z7G
 
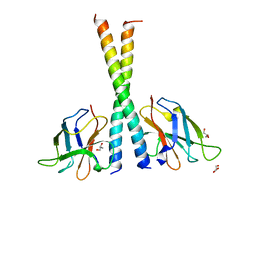 | | Crystal structure of TAX1BP1 SKICH region in complex with NAP1 | | Descriptor: | 5-azacytidine-induced protein 2, GLYCEROL, Tax1-binding protein 1 | | Authors: | Pan, L.F, Fu, T, Liu, J.P, Xie, X.Q, Wang, Y.L, Hu, S.C. | | Deposit date: | 2018-01-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YKB
 
 | |
5ZZS
 
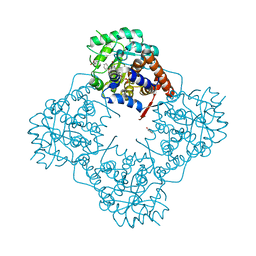 | | The crystal structure of Mandelate oxidase with benzoic acid | | Descriptor: | 4-hydroxymandelate oxidase, BENZOIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-04 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Biochemical and structural explorations of alpha-hydroxyacid oxidases reveal a four-electron oxidative decarboxylation reaction.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
