4GP8
 
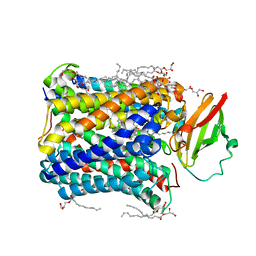 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133W+T231F from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
4EJL
 
 | |
4EJ8
 
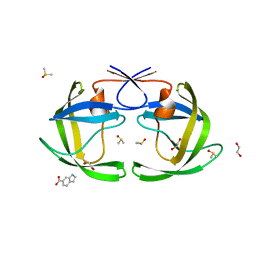 | |
4GQS
 
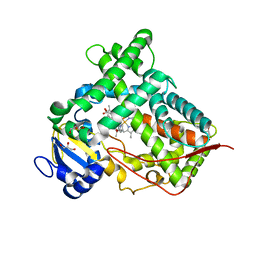 | | Structure of Human Microsomal Cytochrome P450 (CYP) 2C19 | | Descriptor: | (4-hydroxy-3,5-dimethylphenyl)(2-methyl-1-benzofuran-3-yl)methanone, Cytochrome P450 2C19, GLYCEROL, ... | | Authors: | Reynald, R.L, Sansen, S, Stout, C.D, Johnson, E.F. | | Deposit date: | 2012-08-23 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Characterization of Human Cytochrome P450 2C19: ACTIVE SITE DIFFERENCES BETWEEN P450s 2C8, 2C9, AND 2C19.
J.Biol.Chem., 287, 2012
|
|
4GP5
 
 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133W from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
4G71
 
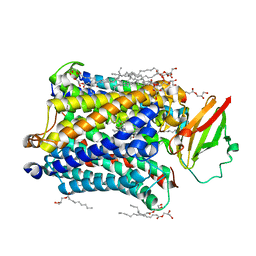 | |
4G7S
 
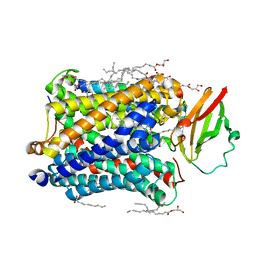 | |
4EJD
 
 | |
1BR3
 
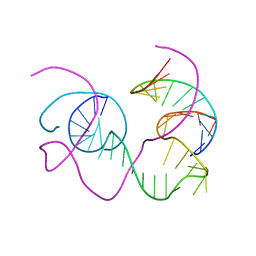 | | CRYSTAL STRUCTURE OF AN 82-NUCLEOTIDE RNA-DNA COMPLEX FORMED BY THE 10-23 DNA ENZYME | | Descriptor: | DNA (10-23 DNA ENZYME), RNA (5'-R(*GP*GP*AP*CP*AP*GP*AP*UP*GP*GP*GP*AP*G)-3') | | Authors: | Nowakowski, J, Shim, P.J, Prasad, G.S, Stout, C.D, Joyce, G.F. | | Deposit date: | 1998-08-13 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10-23 DNA enzyme.
Nat.Struct.Biol., 6, 1999
|
|
1B0J
 
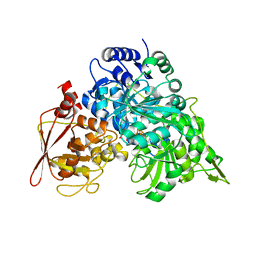 | | CRYSTAL STRUCTURE OF ACONITASE WITH ISOCITRATE | | Descriptor: | ACONITATE HYDRATASE, IRON/SULFUR CLUSTER, ISOCITRIC ACID | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1998-11-10 | | Release date: | 1998-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex
Protein Sci., 8, 1999
|
|
1A8P
 
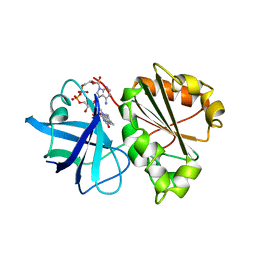 | | FERREDOXIN REDUCTASE FROM AZOTOBACTER VINELANDII | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH:FERREDOXIN OXIDOREDUCTASE | | Authors: | Prasad, G.S, Kresge, N, Muhlberg, A.B, Shaw, A, Jung, Y.S, Burgess, B.K, Stout, C.D. | | Deposit date: | 1998-03-28 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of NADPH:ferredoxin reductase from Azotobacter vinelandii.
Protein Sci., 7, 1998
|
|
1B0T
 
 | | D15K/K84D MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, PROTEIN (FERREDOXIN I) | | Authors: | Sridhar, V, Stout, C.D, Chen, K, Kemper, M.A, Burgess, B.K. | | Deposit date: | 1998-11-12 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the D15K/K84D Mutant of Azotobacter Vinelandii Ferredoxin I
To be Published
|
|
1B0M
 
 | | ACONITASE R644Q:FLUOROCITRATE COMPLEX | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, PROTEIN (ACONITASE) | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1998-11-11 | | Release date: | 1998-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1C96
 
 | | S642A:CITRATE COMPLEX OF ACONITASE | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, MITOCHONDRIAL ACONITASE, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1999-07-31 | | Release date: | 1999-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1B0V
 
 | | I40N MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, PROTEIN (FERREDOXIN) | | Authors: | Sridhar, V, Prasad, G.S, Stout, C.D, Chen, K, Burgess, B.K. | | Deposit date: | 1998-11-12 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Alteration of the reduction potential of the [4Fe-4S](2+/+) cluster of Azotobacter vinelandii ferredoxin I.
J.Biol.Chem., 274, 1999
|
|
1B0K
 
 | | S642A:FLUOROCITRATE COMPLEX OF ACONITASE | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, OXYGEN ATOM, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1C97
 
 | | S642A:ISOCITRATE COMPLEX OF ACONITASE | | Descriptor: | IRON/SULFUR CLUSTER, ISOCITRIC ACID, MITOCHONDRIAL ACONITASE, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1999-07-31 | | Release date: | 1999-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1D4O
 
 | | CRYSTAL STRUCTURE OF TRANSHYDROGENASE DOMAIN III AT 1.2 ANGSTROMS RESOLUTION | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP(H) TRANSHYDROGENASE | | Authors: | Prasad, G.S, Sridhar, V, Yamaguchi, M, Hatefi, Y, Stout, C.D. | | Deposit date: | 1999-10-04 | | Release date: | 2000-01-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal structure of transhydrogenase domain III at 1.2 A resolution.
Nat.Struct.Biol., 6, 1999
|
|
3LYN
 
 | | STRUCTURE OF GREEN ABALONE LYSIN DIMER | | Descriptor: | SPERM LYSIN | | Authors: | Kresge, N, Vacquier, V.D, Stout, C.D. | | Deposit date: | 1999-05-19 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The high resolution crystal structure of green abalone sperm lysin: implications for species-specific binding of the egg receptor.
J.Mol.Biol., 296, 2000
|
|
3K9V
 
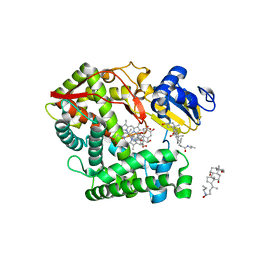 | | Crystal structure of rat mitochondrial P450 24A1 S57D in complex with CHAPS | | Descriptor: | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ... | | Authors: | Annalora, A.J, Goodin, D.B, Hong, W, Zhang, Q, Johnson, E.F, Stout, C.D. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism.
J.Mol.Biol., 396, 2010
|
|
3K9Y
 
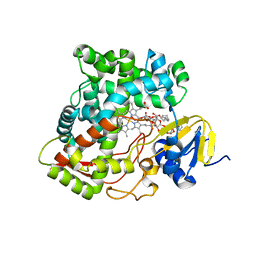 | | Crystal structure of rat mitochondrial P450 24A1 S57D in complex with CYMAL-5 | | Descriptor: | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, ... | | Authors: | Annalora, A.J, Goodin, D.B, Hong, W, Zhang, Q, Johnson, E.F, Stout, C.D. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism.
J.Mol.Biol., 396, 2010
|
|
3QJT
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3QJQ
 
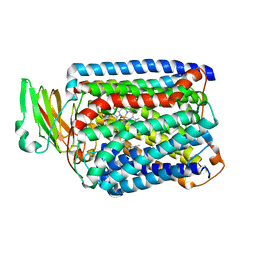 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3QJU
 
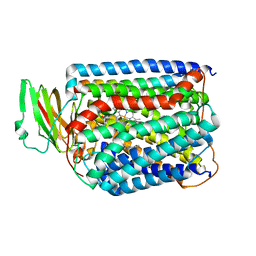 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3QJR
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
