7KKM
 
 | | Structure of the catalytic domain of tankyrase 1 | | Descriptor: | Poly [ADP-ribose] polymerase, ZINC ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
3S2O
 
 | | Fragment based discovery and optimisation of bace-1 inhibitors | | Descriptor: | (3S)-3-(2-amino-5-chloro-1H-benzimidazol-1-yl)-N-[(1R,3S,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-2-yl]pentanamide, Beta-secretase 1, IODIDE ION | | Authors: | Madden, J, Godemann, R, Smith, M.A, Hallett, D, Barker, J, Kraemer, J. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3MSJ
 
 | | Structure of bace (beta secretase) in complex with inhibitor | | Descriptor: | 3-(2-amino-5-chloro-1H-benzimidazol-1-yl)propan-1-ol, BETA-SECRETASE 1, GLYCEROL | | Authors: | Madden, J, Kramer, J, Smith, M.A, Barker, J, Godemann, R. | | Deposit date: | 2010-04-29 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3MSK
 
 | | Fragment Based Discovery and Optimisation of BACE-1 Inhibitors | | Descriptor: | 4-(2-amino-5-chloro-1H-benzimidazol-1-yl)-N-cyclohexyl-N-methylbutanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Smith, M.A, Madden, J.M, Barker, J, Godemann, R, Kraemer, J, Hallett, D. | | Deposit date: | 2010-04-29 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1YCC
 
 | |
1CIF
 
 | |
1CIG
 
 | |
1CIE
 
 | |
1CIH
 
 | |
6XKD
 
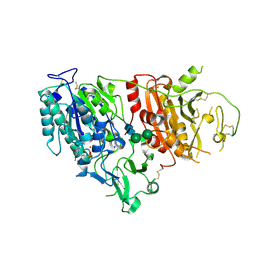 | | Structure of ligand-bound mouse cGAMP hydrolase ENPP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fernandez, D, Li, L. | | Deposit date: | 2020-06-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Aided Development of Small-Molecule Inhibitors of ENPP1, the Extracellular Phosphodiesterase of the Immunotransmitter cGAMP.
Cell Chem Biol, 27, 2020
|
|
6O2E
 
 | |
5I97
 
 | |
6O2F
 
 | |
1AL0
 
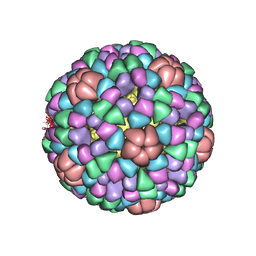 | | PROCAPSID OF BACTERIOPHAGE PHIX174 | | Descriptor: | CAPSID PROTEIN GPF, SCAFFOLDING PROTEIN GPB, SCAFFOLDING PROTEIN GPD, ... | | Authors: | Rossmann, M.G, Dokland, T. | | Deposit date: | 1997-06-06 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a viral procapsid with molecular scaffolding.
Nature, 389, 1997
|
|
2YCC
 
 | |
1RSE
 
 | |
1CD3
 
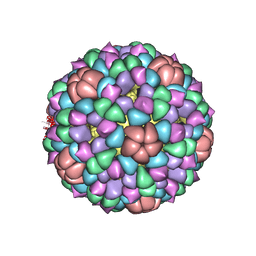 | | PROCAPSID OF BACTERIOPHAGE PHIX174 | | Descriptor: | PROTEIN (CAPSID PROTEIN GPF), PROTEIN (SCAFFOLDING PROTEIN GPB), PROTEIN (SCAFFOLDING PROTEIN GPD), ... | | Authors: | Rossmann, M.G, Dokland, T. | | Deposit date: | 1999-03-05 | | Release date: | 1999-04-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The role of scaffolding proteins in the assembly of the small, single-stranded DNA virus phiX174.
J.Mol.Biol., 288, 1999
|
|
2NTA
 
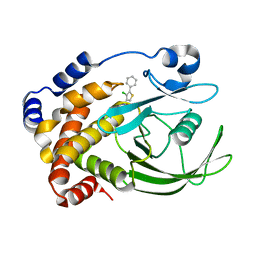 | | Crystal Structure of PTP1B-inhibitor Complex | | Descriptor: | 5-(4-CHLORO-5-PHENYL-3-THIENYL)-1,2,5-THIADIAZOLIDIN-3-ONE 1,1-DIOXIDE, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Follows, B. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing acid replacements of thiophene PTP1B inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2NT7
 
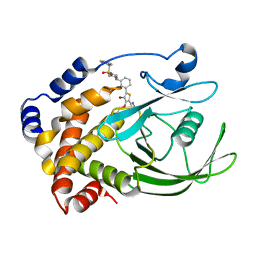 | | Crystal structure of PTP1B-inhibitor complex | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 1, {[5-(3-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}PHENYL)-4-BROMO-2-(2H-TETRAZOL-5-YL)-3-THIENYL]OXY}ACETIC ACID | | Authors: | Xu, W, Follows, B. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing acid replacements of thiophene PTP1B inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1DWT
 
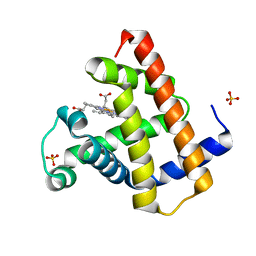 | | Photorelaxed horse heart MYOGLOBIN CO complex | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-12 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
1DWR
 
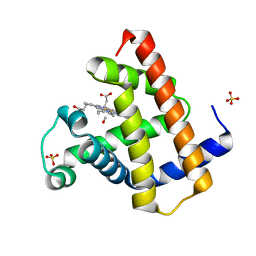 | | MYOGLOBIN (HORSE HEART) WILD-TYPE COMPLEXED WITH CO | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-11 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
1DWS
 
 | | PHOTOLYZED CARBONMONOXY MYOGLOBIN (HORSE HEART) | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-11 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
1GJN
 
 | | Hydrogen Peroxide Derived Myoglobin Compound II at pH 5.2 | | Descriptor: | HYDROXIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hersleth, H.-P, Dalhus, B, Gorbitz, C.H, Andersson, K.K. | | Deposit date: | 2001-07-27 | | Release date: | 2002-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An Iron Hydroxide Moiety in the 1.35 A Resolution Structure of Hydrogen Peroxide Derived Myoglobin Compound II at Ph 5.2
J.Biol.Inorg.Chem., 7, 2002
|
|
3O6Y
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J.K, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-29 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
3NXF
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Jiang, L, Wang, L, Lassila, J.K, Moody, J, Bolduc, J, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-13 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
