6NUL
 
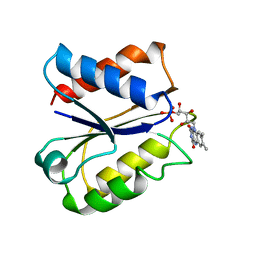 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: N137A REDUCED (150K) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1997-01-09 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
6MST
 
 | | Cryo-EM structure of human AA amyloid fibril | | Descriptor: | Serum amyloid A-1 protein | | Authors: | Loerch, S, Rennegarbe, M, Liberta, F, Grigorieff, N, Fandrich, M, Schmidt, M. | | Deposit date: | 2018-10-18 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids.
Nat Commun, 10, 2019
|
|
5FOH
 
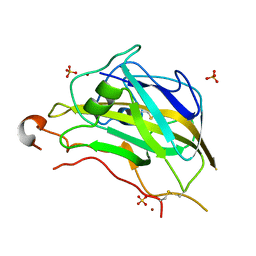 | | Crystal structure of the catalytic domain of NcLPMO9A | | Descriptor: | COPPER (II) ION, LITHIUM ION, POLYSACCHARIDE MONOOXYGENASE, ... | | Authors: | Westereng, B, Kracun, S.K, Dimarogona, M, Mathiesen, G, Willats, W.G.T, Sandgren, M, Aachmann, F.L, Eijsink, V.G.H. | | Deposit date: | 2015-11-18 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparison of three seemingly similar lytic polysaccharide monooxygenases fromNeurospora crassasuggests different roles in plant biomass degradation.
J.Biol.Chem., 2019
|
|
5NNS
 
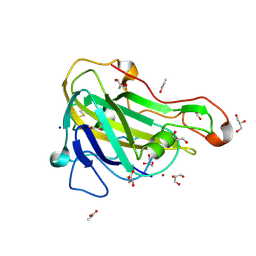 | | Crystal structure of HiLPMO9B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, COPPER (II) ION, ... | | Authors: | Dimarogona, M, Sandgren, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition.
FEBS J., 285, 2018
|
|
6OT3
 
 | | RF2 accommodated state bound Release complex 70S at 24 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6ORL
 
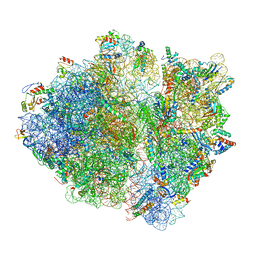 | | RF1 pre-accommodated 70S complex at 24 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OST
 
 | | RF2 pre-accommodated state bound Release complex 70S at 24ms | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
5NLL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN: OXIDIZED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-23 | | Release date: | 1997-03-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
8PX5
 
 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe., solved at wavelength 2.75 A | | Descriptor: | Rpb7-binding protein seb1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Wittmann, S, Renner, M, Grimes, J.M, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
6OUO
 
 | | RF2 accommodated state bound 70S complex at long incubation time | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSQ
 
 | | RF1 accommodated state bound Release complex 70S at long incubation time point | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-26 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSK
 
 | | RF1 accommodated 70S complex at 60 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-01 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
8OG5
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG8
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-methyl-1-phenyl-propan-2-yl)imidazo[2,1-a]isoquinoline, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG6
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 1 | | Descriptor: | 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, ACETATE ION, DDB1- and CUL4-associated factor 1 | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG9
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 4 | | Descriptor: | 5-[1-(4-chlorophenyl)cyclopropyl]imidazo[2,1-a]isoquinoline, DDB1- and CUL4-associated factor 1 | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.945 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OGB
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, DDB1- and CUL4-associated factor 1, DIMETHYL SULFOXIDE, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OGC
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 11 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-(2-azanylethylamino)-2-[1-(4-chlorophenyl)cyclopentyl]quinazolin-7-yl]piperazin-1-yl]ethanone, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG7
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 2 | | Descriptor: | (4~{R})-4-[3-(4-chloranylphenoxy)phenyl]pyrrolidin-2-imine, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OGA
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 6 | | Descriptor: | 1,2-ETHANEDIOL, 5-[1-(4-methoxyphenyl)cyclopropyl]-8-(4-methylpiperazin-1-yl)-2,3-dihydroimidazo[2,1-a]isoquinoline, DDB1- and CUL4-associated factor 1, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
7UYU
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 30 | | Descriptor: | 2-(2,6-difluorophenyl)-4-[4-(pyrrolidine-1-carbonyl)anilino]-5H-pyrrolo[3,4-b]pyridin-5-one, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYR
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 12 | | Descriptor: | 2-(4-{[2-(2,6-difluorophenyl)-5-oxo-5H-pyrrolo[3,4-d]pyrimidin-4-yl]amino}phenyl)-N-ethylacetamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYT
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYW
 
 | | Crystal structure of JAK2 kinase domain in complex with compound 30 | | Descriptor: | 2-(2,6-difluorophenyl)-4-[4-(pyrrolidine-1-carbonyl)anilino]-5H-pyrrolo[3,4-b]pyridin-5-one, Tyrosine-protein kinase JAK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYV
 
 | | Crystal structure of JAK3 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, CHLORIDE ION, Tyrosine-protein kinase JAK3 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
