8FD0
 
 | | Crystal structure of bovine rod opsin in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb2, ... | | Authors: | Salom, D, Palczewski, K, Kiser, P.D. | | Deposit date: | 2022-12-01 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structural basis for the allosteric modulation of rhodopsin by nanobody binding to its extracellular domain.
Nat Commun, 14, 2023
|
|
8FCZ
 
 | | Crystal structure of ground-state rhodopsin in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb2, ... | | Authors: | Salom, D, Palczewski, K, Kiser, P.D. | | Deposit date: | 2022-12-01 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the allosteric modulation of rhodopsin by nanobody binding to its extracellular domain.
Nat Commun, 14, 2023
|
|
8FQ7
 
 | | Nanobody with WIW inserted in CDR3 loop to Inhibit Growth of Alzheimer's Tau fibrils | | Descriptor: | GLYCEROL, Nanobody with WIW insert in CDR3 loop to target tau fibrils | | Authors: | Abskharon, R, Sawaya, M.R, Cascio, D.C, Eisenberg, D.S. | | Deposit date: | 2023-01-05 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based design of nanobodies that inhibit seeding of Alzheimer's patient-extracted tau fibrils.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5VAK
 
 | | Crystal Structure of Beta-Klotho, Domain 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-klotho, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
5VAN
 
 | | Crystal Structure of Beta-Klotho | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
3K80
 
 | | Structure of essential protein from Trypanosoma brucei | | Descriptor: | Antibody, MP18 RNA editing complex protein | | Authors: | Wu, M. | | Deposit date: | 2009-10-13 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of a key interaction protein from the Trypanosoma brucei editosome in complex with single domain antibodies.
J.Struct.Biol., 174, 2011
|
|
5VAQ
 
 | | Crystal Structure of Beta-Klotho in Complex with FGF21CT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-klotho, Fibroblast growth factor 21, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
3K81
 
 | |
3K7U
 
 | | Structure of essential protein from Trypanosoma brucei | | Descriptor: | Antibody, MP18 RNA editing complex protein | | Authors: | Wu, M. | | Deposit date: | 2009-10-13 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of a key interaction protein from the Trypanosoma brucei editosome in complex with single domain antibodies.
J.Struct.Biol., 174, 2011
|
|
5GXB
 
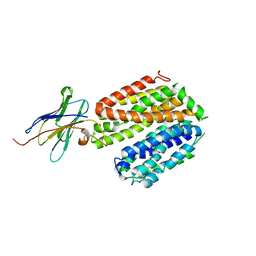 | | crystal structure of a LacY/Nanobody complex | | Descriptor: | Lactose permease, nanobody | | Authors: | Jiang, X, Wu, J.P, Yan, N, Kaback, H.R. | | Deposit date: | 2016-09-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a LacY-nanobody complex in a periplasmic-open conformation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3STB
 
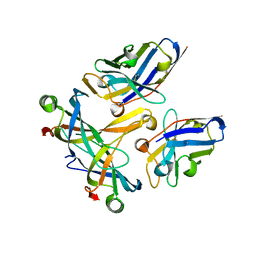 | | A complex of two editosome proteins and two nanobodies | | Descriptor: | MP18 RNA editing complex protein, RNA-editing complex protein MP42, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2011-07-09 | | Release date: | 2011-11-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a heterodimer of editosome interaction proteins in complex with two copies of a cross-reacting nanobody.
Nucleic Acids Res., 40, 2012
|
|
7APJ
 
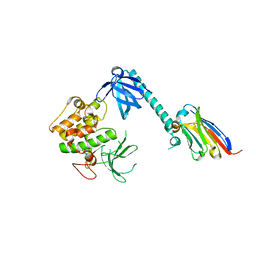 | | Structure of autoinhibited Akt1 reveals mechanism of PIP3-mediated activation | | Descriptor: | NB41, RAC-alpha serine/threonine-protein kinase,Non-specific serine/threonine protein kinase,RAC-alpha serine/threonine-protein kinase | | Authors: | Truebestein, L, Hornegger, H, Leonard, T.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of autoinhibited Akt1 reveals mechanism of PIP 3 -mediated activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5IOF
 
 | |
5G5R
 
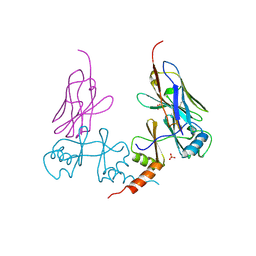 | |
5G5X
 
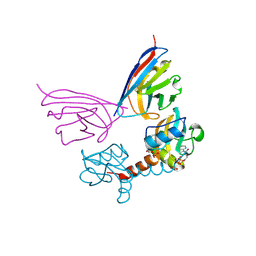 | |
4OCN
 
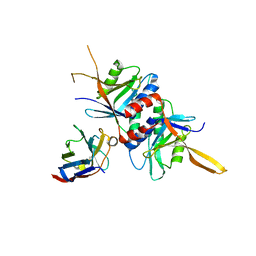 | |
4OCL
 
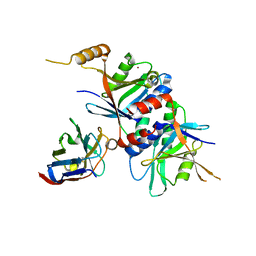 | |
4OCM
 
 | |
6QX3
 
 | | Influenza A virus (A/NT/60/1968) polymerase Hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6RR7
 
 | | Influenza A virus (A/NT/60/1968) polymerase Heterotrimer bound to 3'5' vRNA promoter and capped RNA primer | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(M7G))-R(P*AP*AP*UP*CP*U)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
7RTH
 
 | | Crystal structure of an anti-lysozyme nanobody in complex with an anti-nanobody Fab "NabFab" | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fragment Antigen-Binding Heavy Chain, ... | | Authors: | Filippova, E.V, Mukherjee, S, Bloch, J.S, Locher, K.P, Kossiakoff, A.A. | | Deposit date: | 2021-08-13 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SPA
 
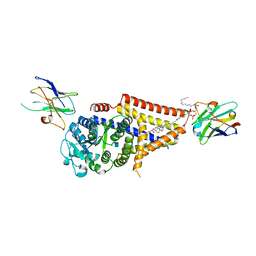 | | Chlorella virus Hyaluronan Synthase in the GlcNAc-primed, channel-open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP8
 
 | | Chlorella virus Hyaluronan Synthase bound to UDP-GlcNAc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP7
 
 | | Chlorella virus hyaluronan synthase inhibited by UDP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP9
 
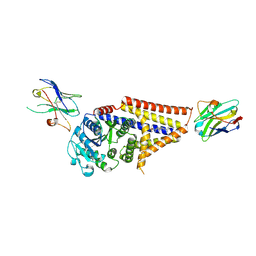 | | Chlorella virus Hyaluronan Synthase in the GlcNAc-primed channel-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
