3PIL
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in reduced form | | Descriptor: | ACETATE ION, Peptide methionine sulfoxide reductase | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3PIM
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in unusual oxidized form | | Descriptor: | Peptide methionine sulfoxide reductase | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3PIN
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in complex with Trx2 | | Descriptor: | Peptide methionine sulfoxide reductase, Thioredoxin-2 | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3OSV
 
 | | The crytsal structure of FLGD from P. Aeruginosa | | Descriptor: | Flagellar basal-body rod modification protein FlgD, GLYCEROL | | Authors: | Wang, D, Luo, M, Niu, S. | | Deposit date: | 2010-09-10 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a novel dimer form of FlgD from P. aeruginosa PAO1
Proteins, 79, 2011
|
|
7VKA
 
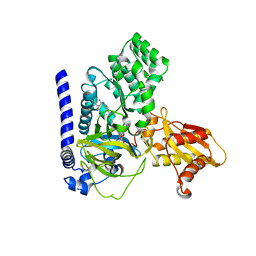 | | Crystal Structure of GH3.6 in complex with an inhibitor | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Indole-3-acetic acid-amido synthetase GH3.6, ... | | Authors: | Wang, N, Luo, M, Bao, H, Huang, H. | | Deposit date: | 2021-09-29 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Chemical genetic screening identifies nalacin as an inhibitor of GH3 amido synthetase for auxin conjugation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UWS
 
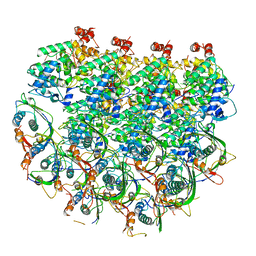 | | Atomic model of the partial VSV nucleocapsid | | Descriptor: | Matrix protein, Nucleoprotein, RNA | | Authors: | Zhou, K, Si, Z, Ge, P, Tsao, J, Luo, M, Zhou, Z.H. | | Deposit date: | 2022-05-03 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Atomic model of vesicular stomatitis virus and mechanism of assembly.
Nat Commun, 13, 2022
|
|
7CUX
 
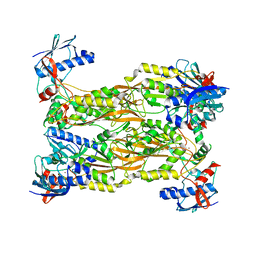 | | Crystal structure of human Schlafen 5 N'-terminal domain (SLFN5-N) involved in ssRNA cleaving and DNA binding | | Descriptor: | Schlafen family member 5, ZINC ION | | Authors: | Yang, J.Y, Luo, M, Ou, J.Y, Wang, Z.W, Gao, S. | | Deposit date: | 2020-08-25 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.29477072 Å) | | Cite: | Crystal structure of human Schlafen 5 N'-terminal domain (SLFN5-N) involved in ssRNA cleaving and DNA binding
To Be Published
|
|
3SLI
 
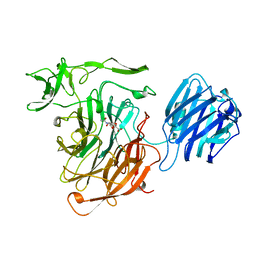 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH 2,7-ANHYDRO-NEU5AC PREPARED BY SOAKING WITH 3'-SIALYLLACTOSE | | Descriptor: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Li, Y.T, Luo, M. | | Deposit date: | 1998-10-03 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A structures of leech intramolecular trans-sialidase complexes: evidence of its enzymatic mechanism.
J.Mol.Biol., 285, 1999
|
|
5WOP
 
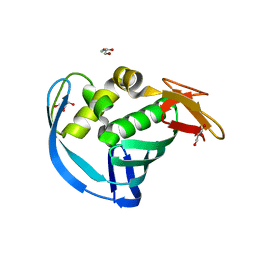 | | High Resolution Structure of Mutant CA09-PB2cap | | Descriptor: | GLYCEROL, Polymerase PB2 | | Authors: | Constantinides, A.E, Gumpper, R.H, Severin, C, Luo, M. | | Deposit date: | 2017-08-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | High-resolution structure of the Influenza A virus PB2cap binding domain illuminates the changes induced by ligand binding.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3CRL
 
 | | Crystal structure of the PDHK2-L2 complex. | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial, MAGNESIUM ION, ... | | Authors: | Popov, K.M, Luo, M, Green, T.J, Grigorian, A, Klyuyeva, A, Tuganova, A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and functional insights into the molecular mechanisms responsible for the regulation of pyruvate dehydrogenase kinase 2.
J.Biol.Chem., 283, 2008
|
|
3ZL9
 
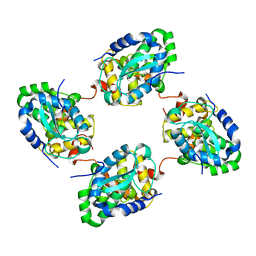 | | Crystal structure of the nucleocapsid protein from Schmallenberg virus | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
3ZLA
 
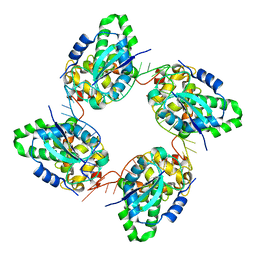 | | Crystal structure of the nucleocapsid protein from Bunyamwera virus bound to RNA | | Descriptor: | NUCLEOPROTEIN, RNA | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
2WYY
 
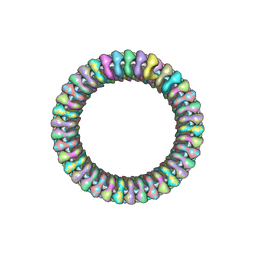 | | CRYOEM MODEL OF THE VESICULAR STOMATITIS VIRUS | | Descriptor: | NUCLEOPROTEIN, POLY-URIDINE | | Authors: | Ge, P, Tsao, J, Green, T.J, Luo, M, Zhou, Z.H. | | Deposit date: | 2009-11-20 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Cryo-Em Model of the Bullet-Shaped Vesicular Stomatitis Virus.
Science, 327, 2010
|
|
4IKP
 
 | | Crystal structure of coactivator-associated arginine methyltransferase 1 with methylenesinefungin | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Dombrovski, L, He, H, Ibanez, G, Wernimont, A, Zheng, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-27 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
5U0A
 
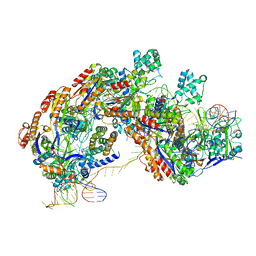 | | CRISPR RNA-guided surveillance complex | | Descriptor: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | Authors: | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
5U07
 
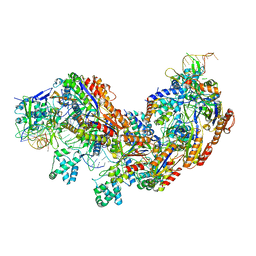 | | CRISPR RNA-guided surveillance complex | | Descriptor: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | Authors: | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
8IDC
 
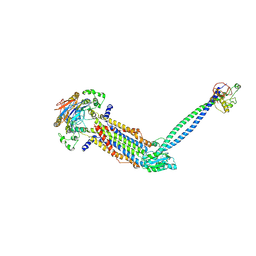 | |
8IGQ
 
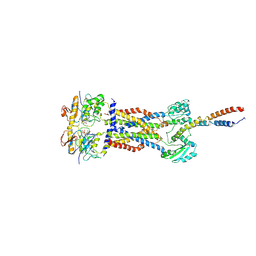 | | Cryo-EM structure of Mycobacterium tuberculosis ADP bound FtsEX/RipC complex in peptidisc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, Luo, M. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Regulation of the cell division hydrolase RipC by the FtsEX system in Mycobacterium tuberculosis.
Nat Commun, 14, 2023
|
|
8IDD
 
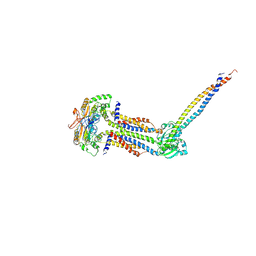 | | Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsEX/RipC complex in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, Luo, M. | | Deposit date: | 2023-02-12 | | Release date: | 2023-10-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Regulation of the cell division hydrolase RipC by the FtsEX system in Mycobacterium tuberculosis.
Nat Commun, 14, 2023
|
|
8IDB
 
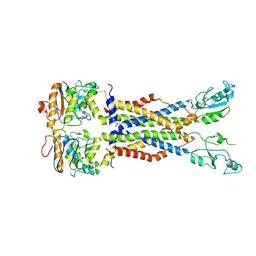 | |
8JIA
 
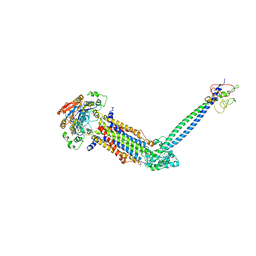 | | Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsE(E165Q)X/RipC complex in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, Luo, M. | | Deposit date: | 2023-05-26 | | Release date: | 2023-10-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Regulation of the cell division hydrolase RipC by the FtsEX system in Mycobacterium tuberculosis.
Nat Commun, 14, 2023
|
|
5WOG
 
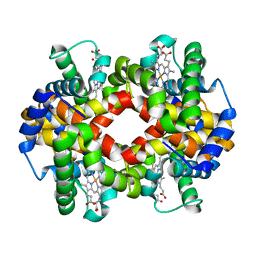 | |
5WOH
 
 | |
9KVM
 
 | | Neutron and X-ray joint refined structure of a copper-containing nitrite reductase (C135A mutant) in complex with formate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, FORMIC ACID | | Authors: | Fukuda, Y, Lintuluoto, M, Hirano, Y, Kusaka, K, Inoue, T, Tamada, T. | | Deposit date: | 2024-12-05 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-02 | | Method: | NEUTRON DIFFRACTION (1.2 Å), X-RAY DIFFRACTION | | Cite: | Structural basis of cuproenzyme nitrite reduction at the level of a single hydrogen atom.
J.Biol.Chem., 301, 2025
|
|
9KWT
 
 | | Structure of a copper-containing nitrite reductase (D98N/G136A mutant) from Geobacillus thermodenitrificans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Lintuluoto, M, Hirano, Y, Kusaka, K, Inoue, T, Tamada, T. | | Deposit date: | 2024-12-06 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Structural basis of cuproenzyme nitrite reduction at the level of a single hydrogen atom.
J.Biol.Chem., 301, 2025
|
|
