2ISW
 
 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with phosphoglycolohydroxamate | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2006-10-18 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization, kinetics, and crystal structures of fructose-1,6-bisphosphate aldolase from the human parasite, Giardia lamblia.
J.Biol.Chem., 282, 2007
|
|
4GAH
 
 | | Human acyl-CoA thioesterases 4 in complex with undecan-2-one-CoA inhibitor | | Descriptor: | Thioesterase superfamily member 4, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3R)-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-4-[[3-oxidanylidene-3-[2-[(2R)-2-oxidanylundecyl]sulfanylethylamino]propyl]amino]butyl] hydrogen phosphate | | Authors: | Lim, K, Pathak, M.C, Herzberg, O. | | Deposit date: | 2012-07-25 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Correlation of structure and function in the human hotdog-fold enzyme hTHEM4.
Biochemistry, 51, 2012
|
|
1DIK
 
 | | PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | PYRUVATE PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Herzberg, O, Chen, C.C.H. | | Deposit date: | 1995-12-06 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
3F5O
 
 | | Crystal Structure of hTHEM2(undecan-2-one-CoA) complex | | Descriptor: | CHLORIDE ION, COENZYME A, HEXAETHYLENE GLYCOL, ... | | Authors: | Xu, H, Gong, W. | | Deposit date: | 2008-11-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mechanisms of human hotdog-fold thioesterase 2 (hTHEM2) substrate recognition and catalysis illuminated by a structure and function based analysis
Biochemistry, 48, 2009
|
|
2FM4
 
 | |
6UPD
 
 | | Structure of trehalose-6-phosphate phosphatase from Salmonella typhimurium in complex with trehalose | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Trehalose-phosphate phosphatase, ... | | Authors: | Harvey, C.M, O'Toole, K.H, Allen, K.N. | | Deposit date: | 2019-10-17 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Structural Analysis of Binding Determinants ofSalmonella typhimuriumTrehalose-6-phosphate Phosphatase Using Ground-State Complexes.
Biochemistry, 59, 2020
|
|
6UPE
 
 | |
6UPC
 
 | |
6UPB
 
 | |
5E0O
 
 | |
3B8I
 
 | |
3L8E
 
 | | Crystal Structure of apo form of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli | | Descriptor: | ACETIC ACID, D,D-heptose 1,7-bisphosphate phosphatase, ZINC ION | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
3L8G
 
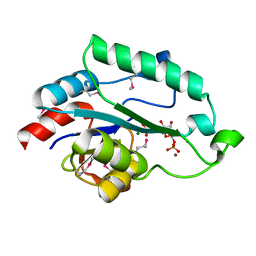 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli complexed with D-glycero-D-manno-heptose 1 ,7-bisphosphate | | Descriptor: | 1,7-di-O-phosphono-L-glycero-beta-D-manno-heptopyranose, D,D-heptose 1,7-bisphosphate phosphatase, MAGNESIUM ION, ... | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
3L8H
 
 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from B. bronchiseptica complexed with magnesium and phosphate | | Descriptor: | FORMIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
2FS2
 
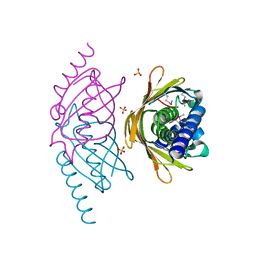 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein paaI, SULFATE ION | | Authors: | Kniewel, R, Buglino, J.A, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, Function, and Mechanism of the Phenylacetate Pathway Hot Dog-fold Thioesterase PaaI
J.Biol.Chem., 281, 2006
|
|
2F0X
 
 | | Crystal structure and function of human thioesterase superfamily member 2(THEM2) | | Descriptor: | SULFATE ION, Thioesterase superfamily member 2 | | Authors: | Cheng, Z, Song, F, Shan, X, Wang, Y, Wei, Z, Gong, W. | | Deposit date: | 2005-11-14 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human thioesterase superfamily member 2
Biochem.Biophys.Res.Commun., 349, 2006
|
|
2DUA
 
 | |
2HJP
 
 | |
2HRW
 
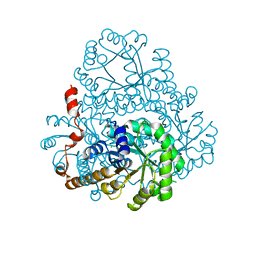 | | Crystal Structure of Phosphonopyruvate Hydrolase | | Descriptor: | CHLORIDE ION, Phosphonopyruvate hydrolase, SODIUM ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Kinetics of Phosphonopyruvate Hydrolase from Voriovorax sp. Pal2: New Insight into the Divergence of Catalysis within the PEP Mutase/Isocitrate Lyase Superfamily
Biochemistry, 45, 2006
|
|
3KZF
 
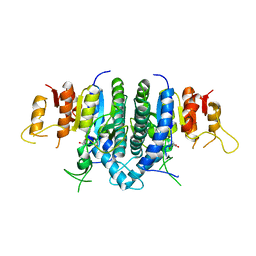 | | Structure of Giardia Carbamate Kinase | | Descriptor: | Carbamate kinase, GLYCEROL | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2009-12-08 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure and characterization of carbamate kinase from the human parasite Giardia lamblia.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3L8F
 
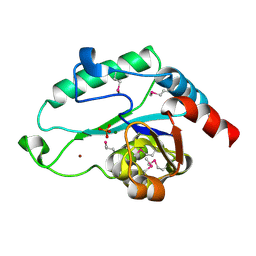 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli complexed with magnesium and phosphate | | Descriptor: | D,D-heptose 1,7-bisphosphate phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
3MMZ
 
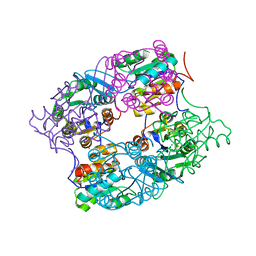 | | CRYSTAL STRUCTURE OF putative HAD family hydrolase from Streptomyces avermitilis MA-4680 | | Descriptor: | CALCIUM ION, CHLORIDE ION, putative HAD family hydrolase | | Authors: | Malashkevich, V.N, Ramagopal, U.A, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3MN1
 
 | | Crystal structure of probable yrbi family phosphatase from pseudomonas syringae pv.phaseolica 1448a | | Descriptor: | CHLORIDE ION, probable yrbi family phosphatase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3N07
 
 | | Structure of putative 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Vibrio cholerae | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, MAGNESIUM ION | | Authors: | Liu, W, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-08-04 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3FA3
 
 | | Crystal structure of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member, trigonal crystal form | | Descriptor: | 2,2-difluoro-3,3-dihydroxybutanedioic acid, 2,3-dimethylmalate lyase, GLYCEROL, ... | | Authors: | Narayanan, B.C, Herzberg, O. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member.
J.Mol.Biol., 386, 2009
|
|
