3S6J
 
 | | The crystal structure of a hydrolase from Pseudomonas syringae | | Descriptor: | CALCIUM ION, Hydrolase, haloacid dehalogenase-like family | | Authors: | Zhang, Z, Syed Ibrahim, B, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-05-25 | | Release date: | 2011-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | The crystal structure of a hydrolase from Pseudomonas syringae
To be Published
|
|
3R03
 
 | |
3R09
 
 | | Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg | | Descriptor: | Hydrolase, haloacid dehalogenase-like family, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-03-07 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg
To be Published
|
|
3T81
 
 | |
3T8L
 
 | | Crystal Structure of adenine deaminase with Mn/Fe | | Descriptor: | Adenine deaminase 2, UNKNOWN ATOM OR ION | | Authors: | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-08-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The catalase activity of diiron adenine deaminase.
Protein Sci., 20, 2011
|
|
3SLR
 
 | | Crystal structure of N-terminal part of the protein BF1531 from Bacteroides fragilis containing phosphatase domain complexed with Mg. | | Descriptor: | MAGNESIUM ION, uncharacterized protein BF1531 | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-20 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Crystal structure of N-terminal part of the protein BF1531 from Bacteroides fragilis
containing phosphatase domain complexed with Mg.
TO BE PUBLISHED
|
|
3SMD
 
 | |
3SN4
 
 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Toro, R, Dickey, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and alpha-ketoglutarate
To be Published
|
|
3SN0
 
 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and fumarate | | Descriptor: | CHLORIDE ION, FUMARIC ACID, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Toro, R, Dickey, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and fumarate
To be Published
|
|
3QKC
 
 | | CRYSTAL STRUCTURE OF geranyl diphosphate synthase small subunit from Antirrhinum majus | | Descriptor: | Geranyl diphosphate synthase small subunit | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of geranyl diphosphate synthase small subunit from Antirrhinum majus
To be Published
|
|
3QDK
 
 | |
3RAZ
 
 | |
3SN1
 
 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and tartrate | | Descriptor: | CHLORIDE ION, D(-)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Toro, R, Dickey, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and tartrate
To be Published
|
|
3TVI
 
 | | Crystal structure of Clostridium acetobutylicum aspartate kinase (CaAK): An important allosteric enzyme for industrial amino acids production | | Descriptor: | ASPARTIC ACID, Aspartokinase, LYSINE | | Authors: | Manjasetty, B.A, Chance, M.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-09-20 | | Release date: | 2011-11-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Clostridium acetobutylicum Aspartate kinase (CaAK): An important allosteric enzyme for amino acids production.
Biotechnol Rep (Amst), 3, 2014
|
|
1UBD
 
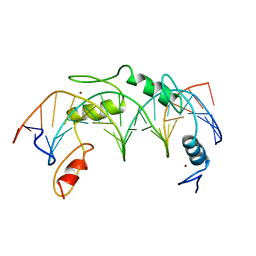 | | CO-CRYSTAL STRUCTURE OF HUMAN YY1 ZINC FINGER DOMAIN BOUND TO THE ADENO-ASSOCIATED VIRUS P5 INITIATOR ELEMENT | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*CP*TP*CP*CP*AP*TP*TP*TP*TP*GP*AP*A P*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*CP*AP*AP*AP*AP*TP*GP*GP*AP*GP*AP*C P*CP*CP*T)-3'), PROTEIN (YY1 ZINC FINGER DOMAIN), ... | | Authors: | Houbaviy, H.B, Usheva, A, Shenk, T, Burley, S.K. | | Deposit date: | 1996-10-04 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1UG3
 
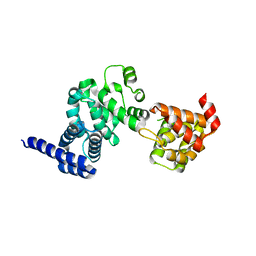 | | C-terminal portion of human eIF4GI | | Descriptor: | eukaryotic protein synthesis initiation factor 4G | | Authors: | Bellsolell, L, Cho-Park, P.F, Poulin, F, Sonenberg, N, Burley, S.K. | | Deposit date: | 2003-06-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1
Structure, 14, 2006
|
|
1UB4
 
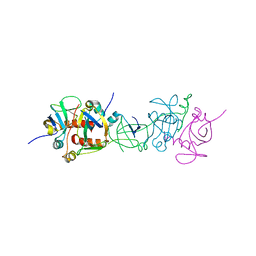 | |
1U02
 
 | | Crystal structure of trehalose-6-phosphate phosphatase related protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-07-20 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications.
Protein Sci., 15, 2006
|
|
1TUF
 
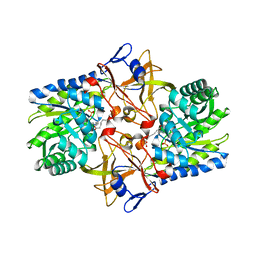 | | Crystal structure of Diaminopimelate Decarboxylase from m. jannaschi | | Descriptor: | AZELAIC ACID, Diaminopimelate decarboxylase | | Authors: | Rajashankar, K, Ray, S.R, Bonanno, J.B, Pinho, M.G, He, G, De Lencastre, H, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-24 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic resistance accessory factor
Structure, 10, 2002
|
|
1TVF
 
 | | Crystal Structure of penicillin-binding protein 4 (PBP4) from Staphylococcus aureus | | Descriptor: | SULFATE ION, UNKNOWN LIGAND, penicillin binding protein 4 | | Authors: | Rajashankar, K.R, Ray, S.S, Bonanno, J.B, Pinho, M, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-29 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of penicillin-binding protein 4 (PBP4) from Staphylococcus aureus
To be Published
|
|
1VOL
 
 | | TFIIB (HUMAN CORE DOMAIN)/TBP (A.THALIANA)/TATA ELEMENT TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*TP*G)-3'), PROTEIN (TATA BINDING PROTEIN (TBP)), ... | | Authors: | Nikolov, D.B, Chen, H, Halay, E.D, Usheva, A.U, Hisatake, K, Lee, D.K, Roeder, R.G, Burley, S.K. | | Deposit date: | 1996-04-29 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a TFIIB-TBP-TATA-element ternary complex.
Nature, 377, 1995
|
|
1VTN
 
 | | CO-CRYSTAL STRUCTURE OF THE HNF-3/FORK HEAD DNA-RECOGNITION MOTIF RESEMBLES HISTONE H5 | | Descriptor: | DNA (5'-D(*GP*AP*CP*TP*AP*AP*GP*TP*CP*AP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*TP*GP*AP*CP*TP*TP*AP*GP*TP*C)-3'), HNF-3/FORK HEAD DNA-RECOGNITION MOTIF, ... | | Authors: | Clark, K.L, Halay, E.D, Lai, E, Burley, S.K. | | Deposit date: | 1995-01-06 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Co-Crystal Structure of the HNF-3/Fork Head DNA-Recognition Motif Resembles Histone H5
Nature, 364, 1993
|
|
1VTO
 
 | |
1VQW
 
 | | Crystal structure of a protein with similarity to flavin-containing monooxygenases and to mammalian dimethylalanine monooxygenases | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN WITH SIMILARITY TO FLAVIN-CONTAINING MONOOXYGENASES AND TO MAMMALIAN DIMETHYLALANINE MONOOXYGENASES | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-05 | | Release date: | 2005-01-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of action of a flavin-containing monooxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1VTL
 
 | |
