5W14
 
 | | ADC-7 in complex with boronic acid transition state inhibitor S03043 | | Descriptor: | 3-{1-[(2R)-2-borono-2-{[(thiophen-2-yl)acetyl]amino}ethyl]-1H-1,2,3-triazol-4-yl}benzoic acid, Beta-lactamase | | Authors: | Smolen, K.A, Powers, R.A, Wallar, B.J. | | Deposit date: | 2017-06-01 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Inhibition of Acinetobacter-Derived Cephalosporinase: Exploring the Carboxylate Recognition Site Using Novel beta-Lactamase Inhibitors.
ACS Infect Dis, 4, 2018
|
|
5WAF
 
 | | ADC-7 in complex with boronic acid transition state inhibitor CR192 | | Descriptor: | Beta-lactamase, phosphonooxy-[[[4-(1~{H}-1,2,3,4-tetrazol-5-yl)-2-(trifluoromethyl)phenyl]sulfonylamino]methyl]borinic acid | | Authors: | Powers, R.A, Wallar, B.J. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Based Analysis of Boronic Acids as Inhibitors of Acinetobacter-Derived Cephalosporinase-7, a Unique Class C beta-Lactamase.
ACS Infect Dis, 4, 2018
|
|
6TZH
 
 | | ADC-7 in complex with boronic acid transition state inhibitor S06015 | | Descriptor: | Beta-lactamase, GLYCINE, PHOSPHATE ION, ... | | Authors: | Fish, E.R, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-06-24 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of beta-Lactamase Inhibitors againstAcinetobacter baumanniiCephalosporinase.
Acs Infect Dis., 6, 2020
|
|
6TZJ
 
 | | ADC-7 in complex with boronic acid transition state inhibitor ME_096 | | Descriptor: | Beta-lactamase, GLYCINE, [4-[[(4-methylphenyl)sulfonylamino]methyl]-1,2,3-triazol-1-yl]methyl-phosphonooxy-borinic acid | | Authors: | Fish, E.R, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of beta-Lactamase Inhibitors againstAcinetobacter baumanniiCephalosporinase.
Acs Infect Dis., 6, 2020
|
|
6TZI
 
 | | ADC-7 in complex with boronic acid transition state inhibitor PFC_001 | | Descriptor: | Beta-lactamase, phosphonooxy-[[4-[[2,2,2-tris(fluoranyl)ethylsulfonylamino]methyl]-1,2,3-triazol-1-yl]methyl]borinic acid | | Authors: | Fish, E.R, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of beta-Lactamase Inhibitors againstAcinetobacter baumanniiCephalosporinase.
Acs Infect Dis., 6, 2020
|
|
6TZF
 
 | | ADC-7 in complex with boronic acid transition state inhibitor S17079 | | Descriptor: | 3-(1-{[hydroxy(phosphonooxy)boranyl]methyl}-1H-1,2,3-triazol-4-yl)benzoic acid, Beta-lactamase | | Authors: | Fish, E.R, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of beta-Lactamase Inhibitors againstAcinetobacter baumanniiCephalosporinase.
Acs Infect Dis., 6, 2020
|
|
6TZG
 
 | | ADC-7 in complex with boronic acid transition state inhibitor S17083 | | Descriptor: | Beta-lactamase, [4-(3-aminocarbonylphenyl)-1,2,3-triazol-1-yl]methyl-phosphonooxy-borinic acid | | Authors: | Fish, E.R, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of beta-Lactamase Inhibitors againstAcinetobacter baumanniiCephalosporinase.
Acs Infect Dis., 6, 2020
|
|
6C6I
 
 | | Crystal structure of a chimeric NDM-1 metallo-beta-lactamase harboring the IMP-1 L3 loop | | Descriptor: | Metallo-beta-lactamase type 2 chimera, ZINC ION | | Authors: | Otero, L, Giannini, E, Klinke, S, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-18 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
6CAC
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase harboring an insertion of a Pro residue in L3 loop | | Descriptor: | CADMIUM ION, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Alzari, P.M, Giannini, E, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-30 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
8CUL
 
 | | Xray ray crystal structure of OXA-24/40 in complex with CR167 | | Descriptor: | 3-({[(dihydroxyboranyl)methyl]sulfamoyl}methyl)benzoic acid, Beta-lactamase | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
8CUQ
 
 | | X-ray crystal structure of ADC-33 in complex with sulfonamidoboronic acid 6e | | Descriptor: | 3-[(4R)-4-ethyl-5,7,7-trihydroxy-2,2,7-trioxo-6-oxa-2lambda~6~-thia-3-aza-7lambda~5~-phospha-5-boraheptan-1-yl]benzoic acid, Beta-lactamase | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
8CUO
 
 | | X-ray crystal structure of OXA-24/40 in complex with sulfonamidoboronic acid 6e | | Descriptor: | 3-({[(1R)-1-boronopropyl]sulfamoyl}methyl)benzoic acid, Beta-lactamase, SULFATE ION | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
8CUP
 
 | | X-ray crystal structure of ADC-33 in complex with sulfonamidoboronic acid 6d | | Descriptor: | 3-[(4S)-4-ethyl-5,7,7-trihydroxy-2,2,7-trioxo-6-oxa-2lambda~6~-thia-3-aza-7lambda~5~-phospha-5-boraheptan-1-yl]benzoic acid, Beta-lactamase | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
8CUM
 
 | | X-ray crystal structure of OXA-24/40 in complex with sulfonamidoboronic acid 6d | | Descriptor: | 3-({[(1S)-1-boronopropyl]sulfamoyl}methyl)benzoic acid, Beta-lactamase, SULFATE ION | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
4X6T
 
 | | M.tuberculosis betalactamase complexed with inhibitor EC19 | | Descriptor: | 3-[(2R)-2-(dihydroxyboranyl)-2-{[(2R)-2-{[(4-ethyl-2,3-dioxo-3,4-dihydropyrazin-1(2H)-yl)carbonyl]amino}-2-(4-hydroxyphenyl)acetyl]amino}ethyl]benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S. | | Deposit date: | 2014-12-09 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibiting the beta-Lactamase of Mycobacterium tuberculosis (Mtb) with Novel Boronic Acid Transition-State Inhibitors (BATSIs).
ACS Infect Dis, 1, 2015
|
|
7KIS
 
 | | Crystal structure of Pseudomonas aeruginosa PBP2 in complex with WCK 5153 | | Descriptor: | (2S,5R)-1-formyl-N'-[(3R)-pyrrolidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, CHLORIDE ION, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Rajavel, M, van den Akker, F. | | Deposit date: | 2020-10-24 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.869 Å) | | Cite: | Structural Characterization of Diazabicyclooctane beta-Lactam "Enhancers" in Complex with Penicillin-Binding Proteins PBP2 and PBP3 of Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7KIV
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F. | | Deposit date: | 2020-10-24 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | Structural Characterization of Diazabicyclooctane beta-Lactam "Enhancers" in Complex with Penicillin-Binding Proteins PBP2 and PBP3 of Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7KIW
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with zidebactam | | Descriptor: | (2S,5R)-1-formyl-N'-[(3R)-piperidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2020-10-25 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Characterization of Diazabicyclooctane beta-Lactam "Enhancers" in Complex with Penicillin-Binding Proteins PBP2 and PBP3 of Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7KIT
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with WCK 4234 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carbonitrile, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F. | | Deposit date: | 2020-10-24 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural Characterization of Diazabicyclooctane beta-Lactam "Enhancers" in Complex with Penicillin-Binding Proteins PBP2 and PBP3 of Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
6OP6
 
 | | Structure of VIM-20 in the reduced state | | Descriptor: | Metallo-beta-lactamase VIM-20, SODIUM ION, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
6OWS
 
 | |
6OP7
 
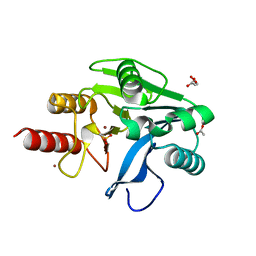 | | Structure of oxidized VIM-20 | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-20, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
4NQ6
 
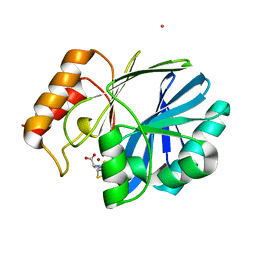 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 complexed with compound L-CS319 | | Descriptor: | (3S,5S,7aR)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Beta-lactamase 2, POTASSIUM ION, ... | | Authors: | Gonzalez, J.M, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2013-11-23 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4NQ2
 
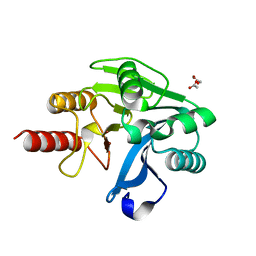 | | Structure of Zn(II)-bound metallo-beta-lactamse VIM-2 from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Aitha, M, Nix, J.C, Crowder, M.W, Page, R.C. | | Deposit date: | 2013-11-23 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical, Mechanistic, and Spectroscopic Characterization of Metallo-beta-lactamase VIM-2.
Biochemistry, 53, 2014
|
|
4QHC
 
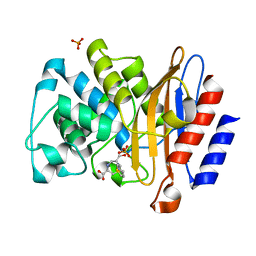 | | Structure of M.Tuberculosis Betalactamase (Blac) with inhibitor having novel mechanism | | Descriptor: | (3R,6R,7S)-7-[(2R,3aR)-hexahydropyrazolo[1,5-c][1,3]thiazin-2-yl]-6-(hydroxymethyl)-1,4-thiazepane-3-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S, Blanchard, J. | | Deposit date: | 2014-05-28 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Kinetic and Structural Characterization of the Interaction of 6-Methylidene Penem 2 with the beta-Lactamase from Mycobacterium tuberculosis.
Biochemistry, 54, 2015
|
|
