1RLV
 
 | |
5U2A
 
 | |
1PVQ
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, LNSGG BOUND TO THE ENGINEERED RECOGNITION SITE LOXM7 | | Descriptor: | DNA 34-MER, Recombinase cre | | Authors: | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | Deposit date: | 2003-06-28 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
1PVR
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, LNSGG BOUND TO THE LOXP (WILDTYPE) RECOGNITION SITE | | Descriptor: | 34-MER, Recombinase CRE | | Authors: | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | Deposit date: | 2003-06-28 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
1PVP
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, ALSHG BOUND TO THE ENGINEERED RECOGNITION SITE LOXM7 | | Descriptor: | 34-MER, Recombinase cre | | Authors: | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | Deposit date: | 2003-06-28 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
4PN3
 
 | | Crystal structure of 3-hydroxyacyl-CoA-dehydrogenase from Brucella melitensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyacyl-CoA dehydrogenase | | Authors: | Lukacs, C.M, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of 3-hydroxyacyl-CoA-dehydrogenase from Brucella melitensis
To Be Published
|
|
8UOJ
 
 | | Crystal structure of human NUAK1-MARK3 kinase domain chimera bound with azepane (R)-#50 small molecule inhibitor | | Descriptor: | (6M)-4-{[(2R)-azepan-2-yl]methyl}-6-[(4R)-imidazo[1,2-a]pyridin-3-yl]-2H-pyrido[3,2-b][1,4]oxazin-3(4H)-one, 1,2-ETHANEDIOL, BENZOIC ACID, ... | | Authors: | Delker, S.L, Abendroth, J, Mayclin, S.J. | | Deposit date: | 2023-10-19 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of UCB9386 a Potent, Selective and Brain-Penetrant Nuak1 Inhibitor suitable for in vivo Pharmacological Studies
To Be Published
|
|
4PBC
 
 | |
4PFZ
 
 | |
2X9P
 
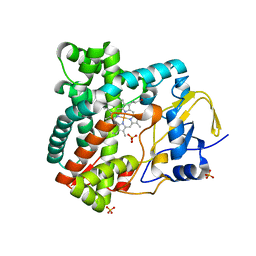 | | X-ray structure of the substrate-free cytochrome P450 PimD - a polyene macrolide antibiotic pimaricin epoxidase | | Descriptor: | PIMD PROTEIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kells, P.M, Ouellet, H, Santos-Aberturas, J, Aparicio, J.F, Podust, L.M. | | Deposit date: | 2010-03-23 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Cytochrome P450 Pimd Suggests Epoxidation of the Polyene Macrolide Pimaricin Occurs Via a Hydroperoxoferric Intermediate.
Chem.Biol., 17, 2010
|
|
2XBK
 
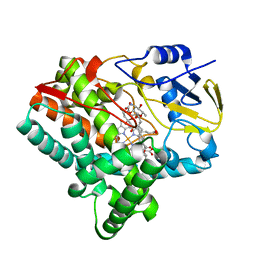 | | X-ray structure of the substrate-bound cytochrome P450 PimD - a polyene macrolide antibiotic pimaricin epoxidase | | Descriptor: | 4,5-DE-EPOXYPIMARICIN, PIMD PROTEIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kells, P.M, Ouellet, H, Santos-Aberturas, J, Aparicio, J.F, Podust, L.M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Cytochrome P450 Pimd Suggests Epoxidation of the Polyene Macrolide Pimaricin Occurs Via a Hydroperoxoferric Intermediate.
Chem.Biol., 17, 2010
|
|
3U0B
 
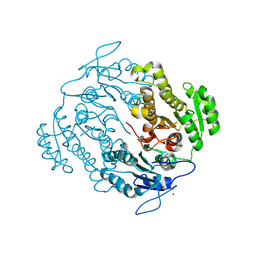 | | Crystal structure of an oxidoreductase from Mycobacterium smegmatis | | Descriptor: | Oxidoreductase, short chain dehydrogenase/reductase family protein, SODIUM ION | | Authors: | Arakaki, T.L, Staker, B.L, Clifton, M.C, Abendroth, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-05 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of FabG4 from Mycolicibacterium smegmatis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
3TE8
 
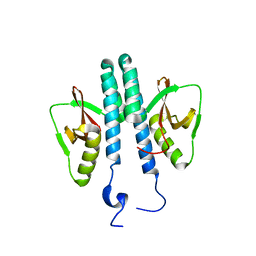 | |
3UWA
 
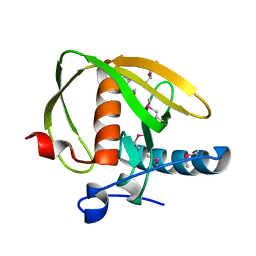 | | Crystal structure of a probable peptide deformylase from synechococcus phage S-SSM7 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, RIIA-RIIB membrane-associated protein, ZINC ION | | Authors: | Lorimer, D, Abendroth, J, Edwards, T.E, Burgin, A, Segall, A, Rohwer, F. | | Deposit date: | 2011-12-01 | | Release date: | 2013-01-09 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
3UWB
 
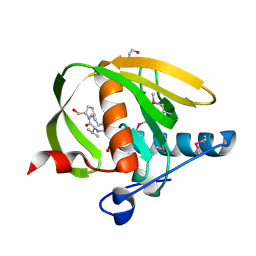 | | Crystal structure of a probable peptide deformylase from strucynechococcus phage S-SSM7 in complex with actinonin | | Descriptor: | 1,2-ETHANEDIOL, ACTINONIN, CHLORIDE ION, ... | | Authors: | Lorimer, D, Abendroth, J, Edwards, T.E, Burgin, A, Segall, A, Rohwer, F. | | Deposit date: | 2011-12-01 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
1KHM
 
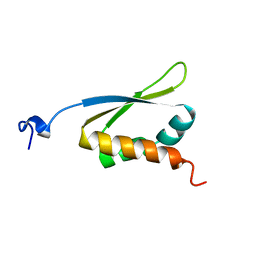 | | C-TERMINAL KH DOMAIN OF HNRNP K (KH3) | | Descriptor: | PROTEIN (HNRNP K) | | Authors: | Baber, J, Libutti, D, Levens, D, Tjandra, N. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor.
J.Mol.Biol., 289, 1999
|
|
3NJB
 
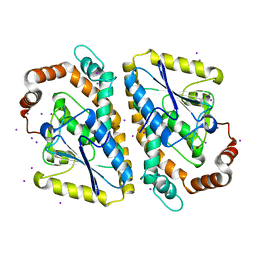 | |
5IZ4
 
 | |
5J6B
 
 | |
4W5K
 
 | |
4LW8
 
 | |
6EFX
 
 | |
6EFW
 
 | |
7SKB
 
 | |
