7ZKZ
 
 | |
7ZKW
 
 | |
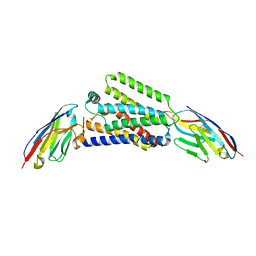
5FMQ
 
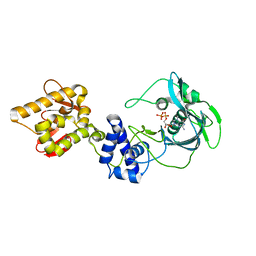 | | Crystal structure of the mid, cap-binding, mid-link and 627 domains from avian influenza A virus polymerase PB2 subunit bound to M7GTP H32 crystal form | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, INFLUENZA A PB2 SUBUNIT | | Authors: | Thierry, E, Pflug, A, Hart, D, Cusack, S. | | Deposit date: | 2015-11-07 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
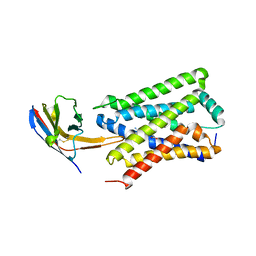
5FML
 
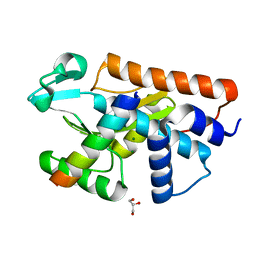 | | Crystal structure of the endonuclease from the PA subunit of influenza B virus bound to the PB2 subunit NLS peptide | | Descriptor: | GLYCEROL, MAGNESIUM ION, PA SUBUNIT OF INFLUENZA B POLYMERASE, ... | | Authors: | Guilligay, D, Gaudon, S, Cusack, S. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
5FMZ
 
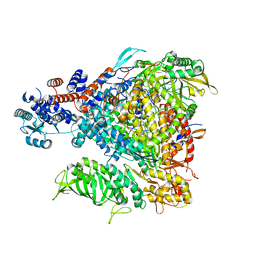 | | Crystal structure of Influenza B polymerase with bound 5' vRNA | | Descriptor: | 5'-R(*AP*GP*UP*AP*GP*UP*AP*AP*CP*AP*AP*GP)-3', POLYMERASE ACIDIC PROTEIN, POLYMERASE BASIC PROTEIN 2, ... | | Authors: | Guilligay, D, Cusack, S. | | Deposit date: | 2015-11-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
5FMM
 
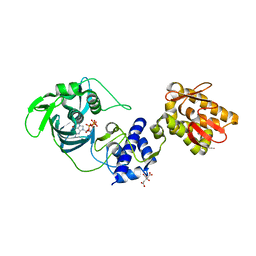 | | crystal structure of the mid, cap-binding, mid-link and 627 domains from avian influenza a virus polymerase PB2 subunit bound to M7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CITRATE ANION, INFLUENZA A PB2 SUBUNIT | | Authors: | Thierry, E, Pflug, A, Hart, D, Cusack, S. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
6EQD
 
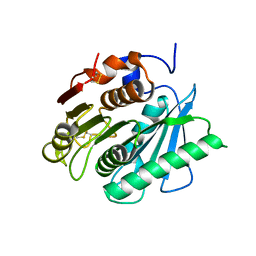 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis collected at long wavelength | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQE
 
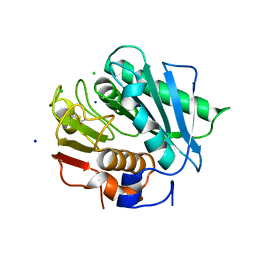 | | High resolution crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SODIUM ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQF
 
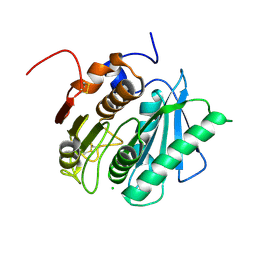 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup P212121 | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7ACV
 
 | |
7ACW
 
 | |
7ACZ
 
 | |
7ACY
 
 | |
8ENF
 
 | |
8END
 
 | |
8ENB
 
 | |
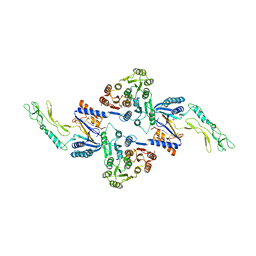
3ZET
 
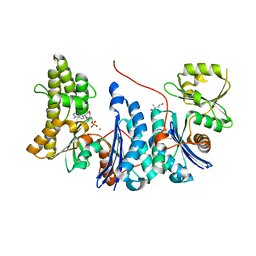 | | Structure of a Salmonella typhimurium YgjD-YeaZ heterodimer. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CADMIUM ION, PROBABLE TRNA THREONYLCARBAMOYLADENOSINE BIOSYNTHESIS PROTEIN GCP, ... | | Authors: | Nichols, C.E, Lamb, H.K, Thompson, P, El Omari, K, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of the Dimer of Two Essential Salmonella Typhimurium Proteins, Ygjd & Yeaz and Calorimetric Evidence for the Formation of a Ternary Ygjd-Yeaz-Yjee Complex.
Protein Sci., 22, 2013
|
|
3ZEU
 
 | | Structure of a Salmonella typhimurium YgjD-YeaZ heterodimer bound to ATPgammaS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Nichols, C.E, Lamb, H.K, Thompson, P, El Omari, K, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-20 | | Last modified: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Crystal Structure of the Dimer of Two Essential Salmonella Typhimurium Proteins, Ygjd & Yeaz and Calorimetric Evidence for the Formation of a Ternary Ygjd-Yeaz-Yjee Complex.
Protein Sci., 22, 2013
|
|
6QNR
 
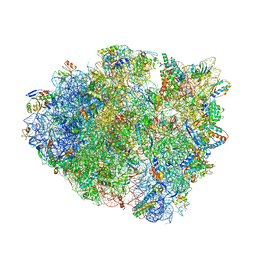 | | 70S ribosome elongation complex (EC) with experimentally assigned potassium ions | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2019-02-11 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction.
Nat Commun, 10, 2019
|
|
6QNQ
 
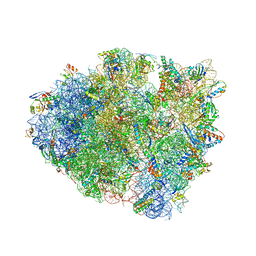 | | 70S ribosome initiation complex (IC) with experimentally assigned potassium ions | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2019-02-11 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction.
Nat Commun, 10, 2019
|
|
5MDT
 
 | |
7QGQ
 
 | |
5EPI
 
 | |
6JJI
 
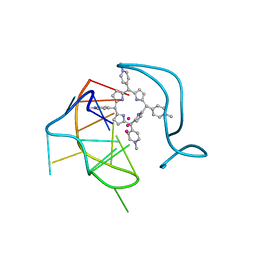 | | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 (1:1) | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, POTASSIUM ION, RNA (5'-R(*GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3') | | Authors: | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
4AEJ
 
 | | Crystal structure of Human fibrillar procollagen type III C- propeptide trimer | | Descriptor: | CALCIUM ION, COLLAGEN ALPHA-1(III) CHAIN, GLYCEROL | | Authors: | Bourhis, J.M, Mariano, N, Zhao, Y, Harlos, K, Jones, E.Y, Moali, C, Aghajari, N, Hulmes, D.J.S. | | Deposit date: | 2012-01-11 | | Release date: | 2012-09-12 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis of Fibrillar Collagen Trimerization and Related Genetic Disorders.
Nat.Struct.Mol.Biol., 19, 2012
|
|
