6V0B
 
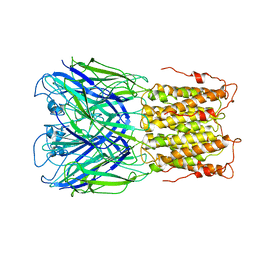 | | Unliganded ELIC in POPC-only nanodiscs. | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Grosman, C, Kumar, P. | | Deposit date: | 2019-11-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4JLQ
 
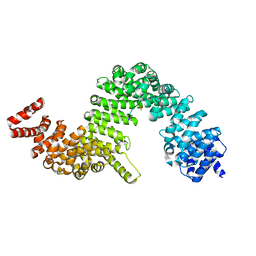 | | Crystal structure of human Karyopherin-beta2 bound to the PY-NLS of Saccharomyces cerevisiae NAB2 | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2, Transportin-1 | | Authors: | Sampathkumar, P, Gizzi, A, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of human Karyopherin beta 2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2.
J.Struct.Funct.Genom., 14, 2013
|
|
6MB5
 
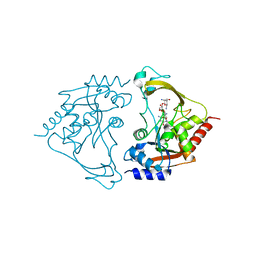 | | AAC-IIIb binary with NEOMYCIN | | Descriptor: | Aac(3)-IIIb protein, NEOMYCIN | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6MB7
 
 | | Binary (paromomycin) structure of AAC-IIIb | | Descriptor: | Aac(3)-IIIb protein, PAROMOMYCIN | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6MB9
 
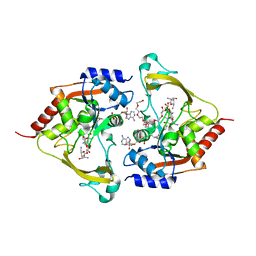 | | Ternary (neomycin/CoA) structure of AAC-IIIb | | Descriptor: | Aac(3)-IIIb protein, COENZYME A, NEOMYCIN, ... | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6MB8
 
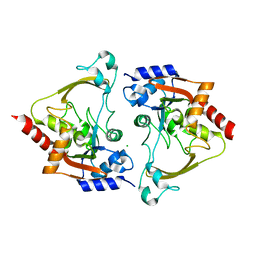 | | Apo structure of AAC-IIIb | | Descriptor: | Aac(3)-IIIb protein, CHLORIDE ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6NTI
 
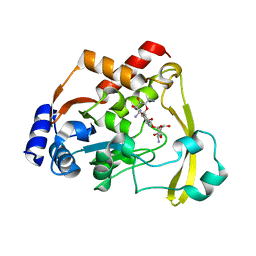 | | Neutron/X-ray crystal structure of AAC-VIa bound to kanamycin b | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.3 Å), X-RAY DIFFRACTION | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6MB4
 
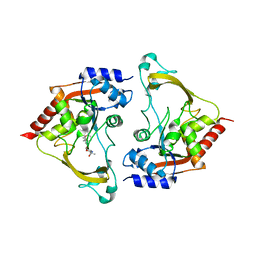 | | Binary (sisomicin) structure of AAC-IIIb | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aac(3)-IIIb protein | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6MB6
 
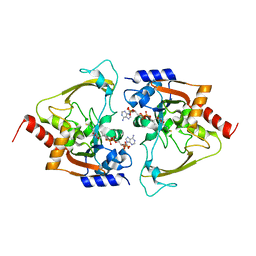 | | AAC-IIIb binary with CoASH | | Descriptor: | Aac(3)-IIIb protein, COENZYME A, MALONATE ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6NTJ
 
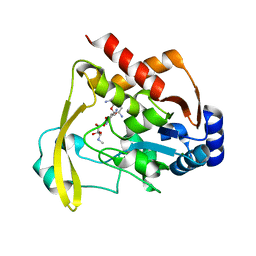 | | Neutron/X-ray crystal structure of AAC-VIa bound to gentamicin C1A | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
2YFJ
 
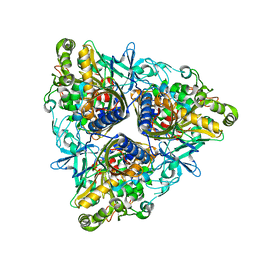 | | Crystal structure of Biphenyl dioxygenase variant RR41 with dibenzofuran | | Descriptor: | BIPHENYL DIOXYGENASE SUBUNIT ALPHA, BIPHENYL DIOXYGENASE SUBUNIT BETA, DIBENZOFURAN, ... | | Authors: | Kumar, P, Sylvestre, M, Bolin, J.T. | | Deposit date: | 2011-04-06 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Retuning Rieske-Type Oxygenases to Expand Substrate Range.
J.Biol.Chem., 286, 2011
|
|
6BBZ
 
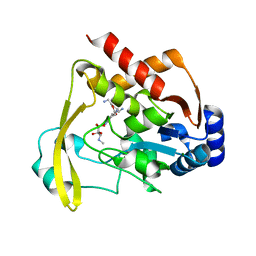 | | Room temperature neutron/X-ray structure of sisomicin bound AAC-VIa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, AAC 3-VI protein, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
6BC4
 
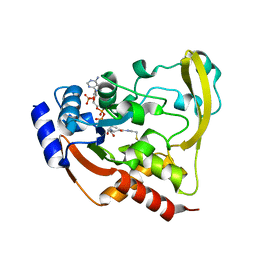 | |
7L6Q
 
 | | Unliganded ELIC in styrene-maleic-acid nanodiscs at 2.5-Angstrom resolution | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CARDIOLIPIN, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Grosman, C, Kumar, P. | | Deposit date: | 2020-12-23 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure and function at the lipid-protein interface of a pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6BBR
 
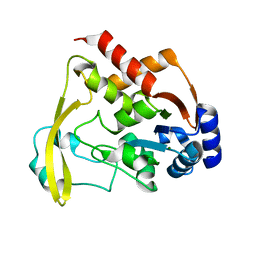 | |
6BC2
 
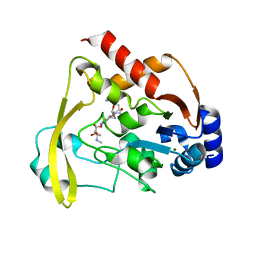 | |
6BC7
 
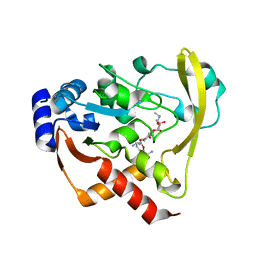 | | Cryo X-ray structure of sisomicin bound AAC-VIa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, AAC 3-VI protein, ACETATE ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
6BC6
 
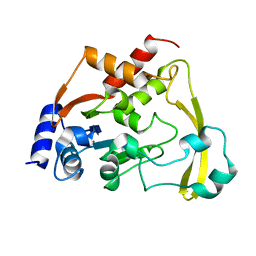 | | Cryo X-ray structure of apo AAC-VIa | | Descriptor: | AAC 3-VI protein, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
6BC3
 
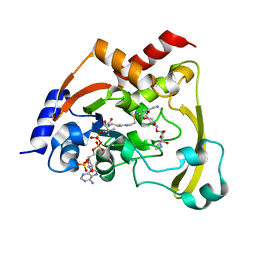 | | Cryo X-ray structure of sisomicin bound AAC-VIa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, AAC 3-VI protein, COENZYME A | | Authors: | Cuneo, M.J, Kumar, P, Serpersu, E.H. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
6BC5
 
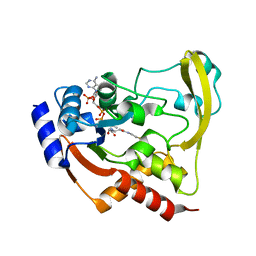 | |
6BMS
 
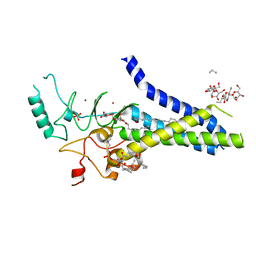 | | Palmitoyltransferase structure | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, DODECYL-BETA-D-MALTOSIDE, PALMITIC ACID, ... | | Authors: | Kumar, P, Rajashankar, K. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
3GZX
 
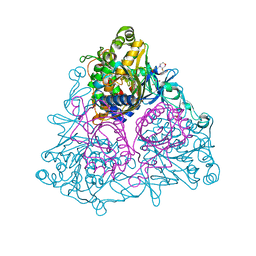 | | Crystal Structure of the Biphenyl Dioxygenase in complex with Biphenyl from Comamonas testosteroni Sp. Strain B-356 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BIPHENYL, Biphenyl dioxygenase subunit alpha, ... | | Authors: | Kumar, P, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2009-04-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Characterization of Pandoraea pnomenusa B-356 Biphenyl Dioxygenase Reveals Features of Potent Polychlorinated Biphenyl-Degrading Enzymes
Plos One, 8, 2013
|
|
3GZY
 
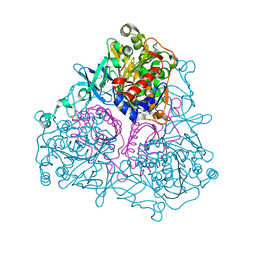 | | Crystal Structure of the Biphenyl Dioxygenase from Comamonas testosteroni Sp. Strain B-356 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Biphenyl dioxygenase subunit alpha, Biphenyl dioxygenase subunit beta, ... | | Authors: | Kumar, P, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2009-04-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Characterization of Pandoraea pnomenusa B-356 Biphenyl Dioxygenase Reveals Features of Potent Polychlorinated Biphenyl-Degrading Enzymes
Plos One, 8, 2013
|
|
7F8P
 
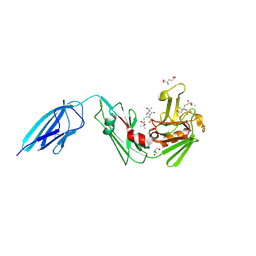 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) with new carbapenem drug T203 | | Descriptor: | (2R,3R)-3-methyl-4-(2-oxidanylidene-2-propan-2-yloxy-ethyl)sulfanyl-2-[(2R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]-2,3-dihydro-1H-pyrrole-5-carboxylic acid, (4R,5S,6S)-6-((R)-1-hydroxyethyl)-3-((2-isopropoxy-2-oxoethyl)thio)-4-methyl-7-oxo-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, GLUTAMIC ACID, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2021-07-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric cooperation in beta-lactam binding to a non-classical transpeptidase.
Elife, 11, 2022
|
|
7F71
 
 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) with peptidoglycan sugar moiety and glutamate | | Descriptor: | GLUTAMIC ACID, GLYCEROL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2021-06-26 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Allosteric cooperation in beta-lactam binding to a non-classical transpeptidase.
Elife, 11, 2022
|
|
