2OCX
 
 | | Crystal structure of Se-Met fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-12-21 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
2NNR
 
 | | Crystal structure of chagasin, cysteine protease inhibitor from Trypanosoma cruzi | | Descriptor: | CHLORIDE ION, Chagasin, GLYCEROL, ... | | Authors: | Redzynia, I, Bujacz, G, Ljunggren, A, Jaskolski, M, Abrahamson, M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease
J.Mol.Biol., 371, 2007
|
|
2NQD
 
 | | Crystal structure of cysteine protease inhibitor, chagasin, in complex with human cathepsin L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Cathepsin L, ... | | Authors: | Redzynia, I, Bujacz, G, Ljunggren, A, Jaskolski, M, Abrahamson, M. | | Deposit date: | 2006-10-31 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease
J.Mol.Biol., 371, 2007
|
|
3CBJ
 
 | | Chagasin-Cathepsin B complex | | Descriptor: | Cathepsin B, Chagasin, PHOSPHATE ION | | Authors: | Redzynia, I, Bujacz, G.D, Abrahamson, M, Ljunggren, A, Jaskolski, M, Mort, J.S. | | Deposit date: | 2008-02-22 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Displacement of the occluding loop by the parasite protein, chagasin, results in efficient inhibition of human cathepsin B.
J.Biol.Chem., 283, 2008
|
|
6SJJ
 
 | | A new modulated crystal structure of ANS complex of St John's wort Hyp-1 protein with 36 protein molecules in the asymmetric unit of the supercell | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-ANILINO-1-NAPHTHALENE SULFONATE, CITRATE ANION, ... | | Authors: | Smietanska, J, Sliwiak, J, Gilski, M, Dauter, Z, Strzalka, R, Wolny, J, Jaskolski, M. | | Deposit date: | 2019-08-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new modulated crystal structure of the ANS complex of the St John's wort Hyp-1 protein with 36 protein molecules in the asymmetric unit of the supercell.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3E1Z
 
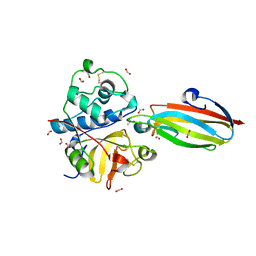 | | Crystal structure of the parasite protesase inhibitor chagasin in complex with papain | | Descriptor: | ACETIC ACID, Chagasin, FORMIC ACID, ... | | Authors: | Redzynia, I, Bujacz, G, Bujacz, A, Ljunggren, A, Abrahamson, M, Jaskolski, M. | | Deposit date: | 2008-08-05 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the parasite inhibitor chagasin in complex with papain allows identification of structural requirements for broad reactivity and specificity determinants for target proteases.
Febs J., 276, 2009
|
|
7ATG
 
 | | Crystal structure of Z-DNA in complex with putrescinium and potassium cations at ultrahigh-resolution | | Descriptor: | 4-azaniumylbutylazanium, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), POTASSIUM ION | | Authors: | Drozdzal, P, Gilski, M, Jaskolski, M. | | Deposit date: | 2020-10-30 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.6 Å) | | Cite: | Crystal structure of Z-DNA in complex with the polyamine putrescine and potassium cations at ultra-high resolution.
Acta Crystallogr.,Sect.B, 77, 2021
|
|
3GAX
 
 | | Crystal structure of monomeric human cystatin C stabilized against aggregation | | Descriptor: | Cystatin-C | | Authors: | Kolodziejczyk, R, Michalska, K, Hernandez-Santoyo, A, Wahlbom, M, Grubb, A, Jaskolski, M. | | Deposit date: | 2009-02-18 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human cystatin C stabilized against amyloid formation.
Febs J., 277, 2010
|
|
1G6X
 
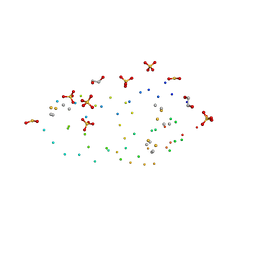 | | ULTRA HIGH RESOLUTION STRUCTURE OF BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) MUTANT WITH ALTERED BINDING LOOP SEQUENCE | | Descriptor: | 1,2-ETHANEDIOL, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Addlagatta, A, Czapinska, H, Krzywda, S, Otlewski, J, Jaskolski, M. | | Deposit date: | 2000-11-08 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Ultrahigh-resolution structure of a BPTI mutant.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3UR8
 
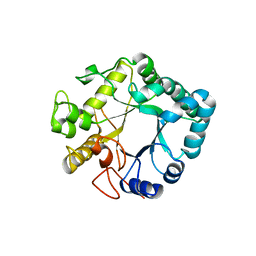 | | Lower-density crystal structure of potato endo-1,3-beta-glucanase | | Descriptor: | Glucan endo-1,3-beta-D-glucosidase | | Authors: | Wojtkowiak, A, Witek, K, Hennig, J, Jaskolski, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Two high-resolution structures of potato endo-1,3-beta-glucanase reveal subdomain flexibility with implications for substrate binding
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6TWT
 
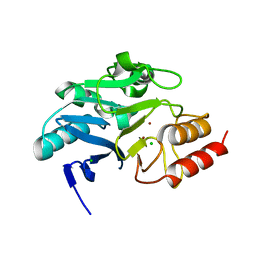 | | Crystal structure of N-terminally truncated NDM-1 metallo-beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Imiolczyk, B, Czyrko-Horczak, J, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
4EID
 
 | | Crystal structure of cytochrome c6 Q57V mutant from Synechococcus sp. PCC 7002 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome c6, HEME C | | Authors: | Krzywda, S, Bialek, W, Zatwarnicki, P, Jaskolski, M, Szczepaniak, A. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Cytochrome c6 and c6C from Synechococcus sp. PCC 7002 - structure and function.
To be Published
|
|
4EIC
 
 | |
2HHC
 
 | | Crystal structure of fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-06-28 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
2HLH
 
 | | Crystal structure of fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | Nodulation fucosyltransferase, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-07-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
1QLQ
 
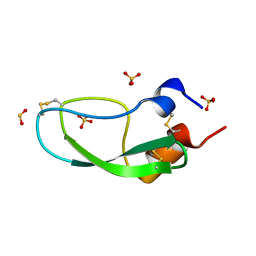 | | Bovine Pancreatic Trypsin Inhibitor (BPTI) Mutant with Altered Binding Loop Sequence | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Czapinska, H, Krzywda, S, Sheldrick, G.M, Otlewski, J, Jaskolski, M. | | Deposit date: | 1999-09-10 | | Release date: | 1999-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High Resolution Structure of Bovine Pancreatic Trypsin Inhibitor with Altered Binding Loop Sequence
J.Mol.Biol., 295, 1999
|
|
3UR7
 
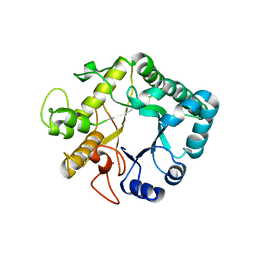 | | Higher-density crystal structure of potato endo-1,3-beta-glucanase | | Descriptor: | Glucan endo-1,3-beta-D-glucosidase, SODIUM ION | | Authors: | Wojtkowiak, A, Witek, K, Hennig, J, Jaskolski, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two high-resolution structures of potato endo-1,3-beta-glucanase reveal subdomain flexibility with implications for substrate binding
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3P4J
 
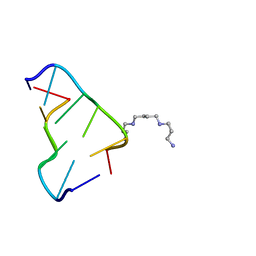 | | Ultra-high resolution structure of d(CGCGCG)2 Z-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SPERMINE | | Authors: | Brzezinski, K, Brzuszkiewicz, A, Dauter, M, Kubicki, M, Jaskolski, M, Dauter, Z. | | Deposit date: | 2010-10-06 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.55 Å) | | Cite: | High regularity of Z-DNA revealed by ultra high-resolution crystal structure at 0.55 A.
Nucleic Acids Res., 39, 2011
|
|
3DR0
 
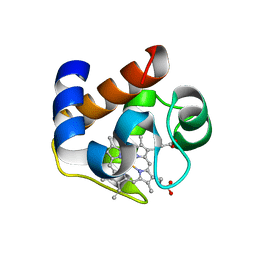 | | Structure of reduced cytochrome c6 from Synechococcus sp. PCC 7002 | | Descriptor: | Cytochrome c6, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Bialek, W, Krzywda, S, Jaskolski, M, Szczepaniak, A. | | Deposit date: | 2008-07-10 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Atomic-resolution structure of reduced cyanobacterial cytochrome c6 with an unusual sequence insertion
Febs J., 276, 2009
|
|
4LUG
 
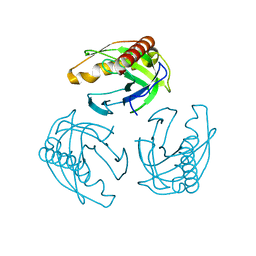 | |
5C9Y
 
 | |
3Q20
 
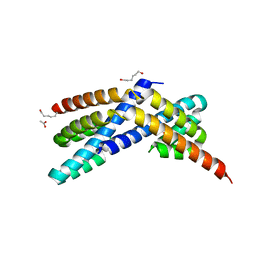 | | Crystal structure of RbcX C103A mutant from Thermosynechococcus elongatus | | Descriptor: | ACETATE ION, HEXANE-1,6-DIOL, RbcX protein | | Authors: | Tarnawski, M, Krzywda, S, Szczepaniak, A, Jaskolski, M. | | Deposit date: | 2010-12-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of the RuBisCO chaperone RbcX from the thermophilic cyanobacterium Thermosynechococcus elongatus
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4HGU
 
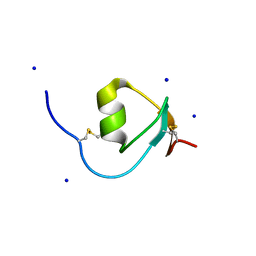 | | Crystal Structure of Galleria mellonella Silk Protease Inhibitor 2 | | Descriptor: | SODIUM ION, Silk protease inhibitor 2 | | Authors: | Krzywda, S, Jaskolski, M, Dvornyk, A, Kludkiewicz, B, Grzelak, K, Zagorski, W, Bal, W, Kopera, E. | | Deposit date: | 2012-10-08 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of a protein prepared by non-enzymatic His-tag removal. Crystallographic and NMR study of GmSPI-2 inhibitor.
Plos One, 9, 2014
|
|
3NX0
 
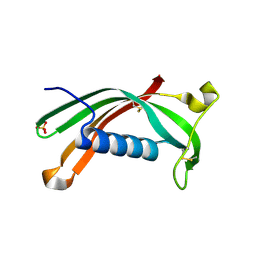 | | Hinge-loop mutation can be used to control 3D domain swapping and amyloidogenesis of human cystatin C | | Descriptor: | Cystatin-C, SULFATE ION | | Authors: | Orlikowska, M, Jankowska, E, Kolodziejczyk, R, Jaskolski, M, Szymanska, A. | | Deposit date: | 2010-07-12 | | Release date: | 2010-12-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Hinge-loop mutation can be used to control 3D domain swapping and amyloidogenesis of human cystatin C.
J.Struct.Biol., 173, 2011
|
|
4PPH
 
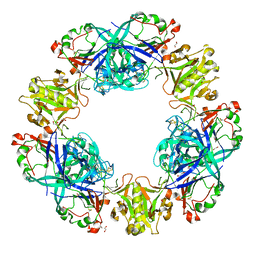 | | Crystal structure of conglutin gamma, a unique basic 7S globulin from lupine seeds | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Czubinski, J, Barciszewski, J, Gilski, M, Lampart-Szczapa, E, Jaskolski, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Structure of gamma-conglutin: insight into the quaternary structure of 7S basic globulins from legumes.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
