3FDZ
 
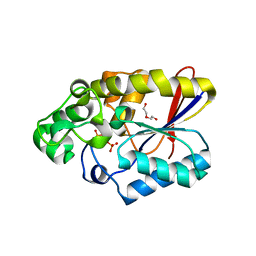 | | Crystal structure of phosphoglyceromutase from burkholderia pseudomallei 1710b with bound 2,3-diphosphoglyceric acid and 3-phosphoglyceric acid | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, 3-PHOSPHOGLYCERIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An ensemble of structures of Burkholderia pseudomallei 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5IF9
 
 | |
5IHP
 
 | |
4PFS
 
 | |
3GW8
 
 | |
3GP5
 
 | |
3GP3
 
 | |
6AUA
 
 | |
6AUB
 
 | |
4MI2
 
 | |
6MJ9
 
 | |
6NB2
 
 | |
6NBM
 
 | |
6CWA
 
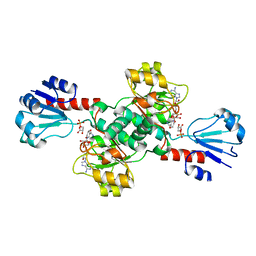 | |
5MW6
 
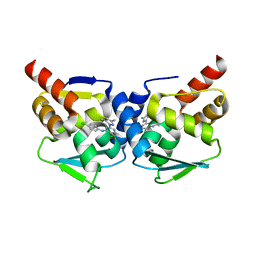 | | Crystal structure of the BCL6 BTB-domain with compound 1 | | Descriptor: | 5-chloranyl-~{N}-(4-chlorophenyl)-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-amine, B-cell lymphoma 6 protein | | Authors: | Davies, D.R, Kessler, D. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
4GNV
 
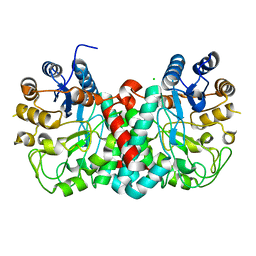 | |
7UMP
 
 | | CRYSTAL STRUCTURE OF PHD2 CATALYTIC DOMAIN (CID 7465) IN COMPLEX WITH AKB-6548 AT 1.8 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Davie, D.R, Abendroth, J, Boyd, J. | | Deposit date: | 2022-04-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Preclinical Characterization of Vadadustat (AKB-6548), an Oral Small Molecule Hypoxia-Inducible Factor Prolyl-4-Hydroxylase Inhibitor, for the Potential Treatment of Renal Anemia.
J.Pharmacol.Exp.Ther., 383, 2022
|
|
3K14
 
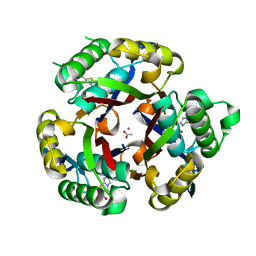 | | Co-crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 535, ethyl 3-methyl-5,6-dihydroimidazo[2,1-b][1,3]thiazole-2-carboxylate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
7S5M
 
 | |
4W65
 
 | |
8DU0
 
 | |
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | Descriptor: | Transforming growth factor beta-2 proprotein | | Authors: | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
4TVI
 
 | |
5VT6
 
 | |
5W8H
 
 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 11 | | Descriptor: | 2-[3-(4-fluorophenyl)-5-(trifluoromethyl)-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lukacs, C.M, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
