1EN7
 
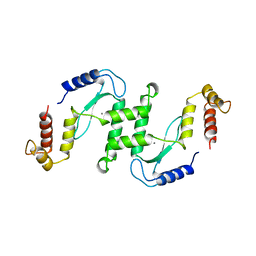 | | ENDONUCLEASE VII (ENDOVII) FROM PHAGE T4 | | Descriptor: | CALCIUM ION, RECOMBINATION ENDONUCLEASE VII, ZINC ION | | Authors: | Raaijmakers, H, Vix, O, Toro, I, Suck, D. | | Deposit date: | 1999-02-07 | | Release date: | 2000-02-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture.
EMBO J., 18, 1999
|
|
3HFA
 
 | |
6RH5
 
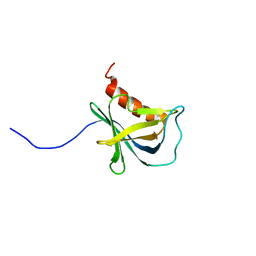 | | Solution structure and 1H, 13C and 15N chemical shift assignments for NECAP1 PHear domain | | Descriptor: | Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
1EJQ
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE | | Descriptor: | SYNDECAN-4 | | Authors: | Shin, J, Oh, E.S, Lee, D, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-04 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE
To be Published
|
|
2MDW
 
 | |
6RKC
 
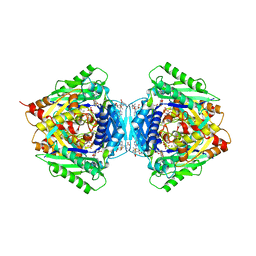 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, Methionine adenosyltransferase, ... | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
1EOE
 
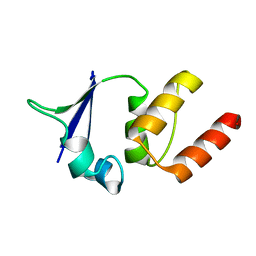 | | CRYSTAL STRUCTURE OF THE V135R MUTANT OF A SHAKER T1 DOMAIN | | Descriptor: | POTASSIUM CHANNEL KV1.1 | | Authors: | Nanao, M.H, Cushman, S.J, Jahng, A.W, DeRubeis, D, Choe, S, Pfaffinger, P.J. | | Deposit date: | 2000-03-22 | | Release date: | 2000-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Voltage dependent activation of potassium channels is coupled to T1 domain structure.
Nat.Struct.Biol., 7, 2000
|
|
1EP0
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-24 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
3EAQ
 
 | |
3EAY
 
 | | Crystal structure of the human SENP7 catalytic domain | | Descriptor: | SULFATE ION, Sentrin-specific protease 7 | | Authors: | Lima, C.D, Reverter, D. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Human SENP7 Catalytic Domain and Poly-SUMO Deconjugation Activities for SENP6 and SENP7.
J.Biol.Chem., 283, 2008
|
|
1EPZ
 
 | | CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOASE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH BOUND LIGAND. | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C, Edwards, A.M. | | Deposit date: | 2000-03-30 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
3HGJ
 
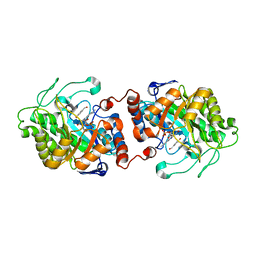 | | Old Yellow Enzyme from Thermus scotoductus SA-01 complexed with p-hydroxy-benzaldehyde | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
2ML7
 
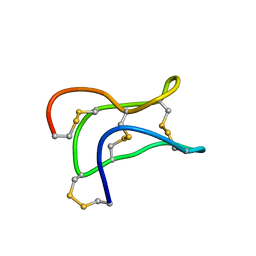 | | Ginsentides: Characterization, Structure and Application of a New Class of Highly Stable Cystine Knot Peptides in Ginseng | | Descriptor: | Specific abundant protein 3 | | Authors: | Wang, S, Nguyen, K, Luo, S, Tam, J, Yang, D. | | Deposit date: | 2014-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ginsentides: Characterization, Structure and Application of a New Class of Highly Stable Cystine Knot Peptides in Ginseng
To be Published
|
|
3V85
 
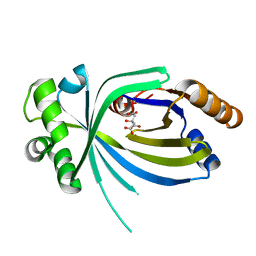 | |
2MBQ
 
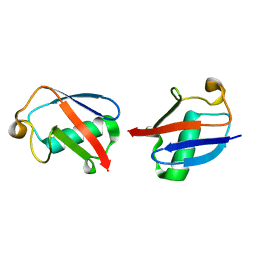 | |
3ES9
 
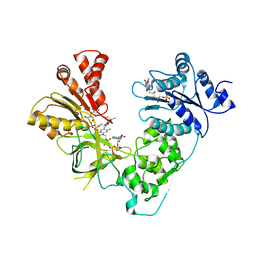 | | NADPH-Cytochrome P450 Reductase in an Open Conformation | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hamdane, D, Xia, C, Im, S.-C, Zhang, H, Kim, J.-J, Waskell, L. | | Deposit date: | 2008-10-05 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450
J.Biol.Chem., 284, 2009
|
|
6RRX
 
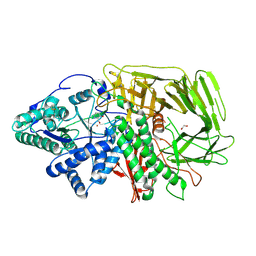 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3R)-2-(Hydroxymethyl)-3-piperidinol | | Descriptor: | (2~{S},3~{R})-2-(hydroxymethyl)piperidin-3-ol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
7YNF
 
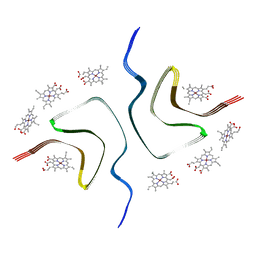 | | CCA-bound alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein, copper;trisodium;18-(2-carboxylatoethyl)-20-(carboxylatomethyl)-12-ethenyl-7-ethyl-3,8,13,17-tetramethyl-17,18-dihydroporphyrin-21,23-diide-2-carboxylate | | Authors: | Tao, Y.Q, Zhao, Q.Y, Liu, C, Li, D. | | Deposit date: | 2022-07-30 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | CCA-bound alpha-synuclein fibrils
To Be Published
|
|
2M0C
 
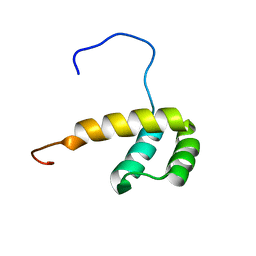 | | Solution NMR Structure of Homeobox Domain of Human ALX4, Northeast Structural Genomics Consortium (NESG) Target HR4490C | | Descriptor: | Homeobox protein aristaless-like 4 | | Authors: | Xu, X, Eletsky, A, Pulavarti, S, Lee, D, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-24 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Homeobox Domain of Human ALX4, Northeast Structural Genomics Consortium (NESG) Target HR4490C
To be Published
|
|
3EUV
 
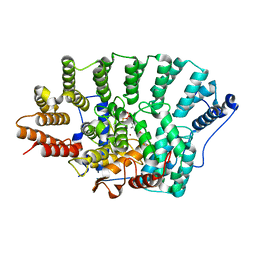 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10, W102T, Y154T) in complex with BiotinGPP | | Descriptor: | (2E,6E)-3,7-dimethyl-8-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)octa-2,6-dien-1-yl trihydrogen diphosphate, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Guo, Z, Nguyen, U.T.T, Delon, C, Bon, R.S, Blankenfeldt, W, Goody, R.S, Waldmann, H, Wolters, D, Alexandrov, K. | | Deposit date: | 2008-10-11 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Analysis of the eukaryotic prenylome by isoprenoid affinity tagging
Nat.Chem.Biol., 5, 2009
|
|
7YQ8
 
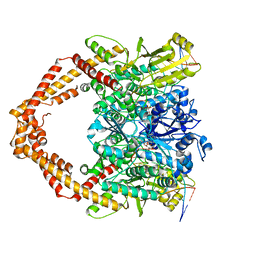 | | Cryo-EM structure of human topoisomerase II beta in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, 50-mer DNA, DNA topoisomerase 2-beta, ... | | Authors: | Naganuma, M, Ehara, H, Kim, D, Nakagawa, R, Cong, A, Bu, H, Jeong, J, Jang, J, Schellenberg, M.J, Bunch, H, Sekine, S. | | Deposit date: | 2022-08-05 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ERK2-topoisomerase II regulatory axis is important for gene activation in immediate early genes.
Nat Commun, 14, 2023
|
|
2MMZ
 
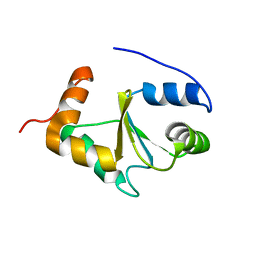 | | Solution structure of the apo form of human glutaredoxin 5 | | Descriptor: | Glutaredoxin-related protein 5, mitochondrial | | Authors: | Banci, L, Brancaccio, D, Ciofi-Baffoni, S, Del Conte, R, Gadepalli, R, Mikolajczyk, M, Neri, S, Piccioli, M, Winkelmann, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | [2Fe-2S] cluster transfer in iron-sulfur protein biogenesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3VDN
 
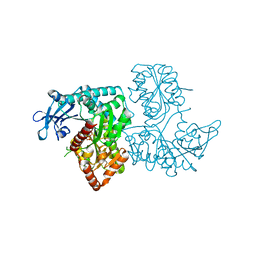 | | Crystal Structure of VldE, the pseudo-glycosyltransferase, in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, VldE | | Authors: | Cavalier, M.C, Yim, Y.-S, Asamizu, S, Neau, D, Mahmud, T, Lee, Y.-H. | | Deposit date: | 2012-01-05 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the VldE, the pseudo-glycosyltransferase, which catalyzes non-glycosidic C-N coupling in Validamycin A biosynthesis
To be Published
|
|
1EJ2
 
 | | Crystal structure of methanobacterium thermoautotrophicum nicotinamide mononucleotide adenylyltransferase with bound NAD+ | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Saridakis, V, Christendat, D, Kimber, M.S, Edwards, A.M, Pai, E.F, Midwest Center for Structural Genomics (MCSG), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-29 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.
J.Biol.Chem., 276, 2001
|
|
7YO2
 
 | |
