6WPG
 
 | | Structural Basis of Salicylic Acid Perception by Arabidopsis NPR Proteins | | Descriptor: | 2-HYDROXYBENZOIC ACID, Regulatory protein NPR4 | | Authors: | Wang, W, Withers, J, Li, H, Zwack, P.J, Rusnac, D.V, Shi, H, Liu, L, Yan, S, Hinds, T.R, Guttman, M, Dong, X, Zheng, N. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.283 Å) | | Cite: | Structural basis of salicylic acid perception by Arabidopsis NPR proteins.
Nature, 586, 2020
|
|
7KZI
 
 | | Intermediate state (QQQ) of near full-length DnaK alternatively fused with a substrate peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Chaperone protein DnaK fused with substrate peptide,Chaperone protein DnaK fused with substrate peptide, ... | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Intermediates in allosteric equilibria of DnaK-ATP interactions with substrate peptides
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7KZU
 
 | | Quasi-intermediate state (Q) of a truncated Hsp70 DnaK fused with a substrate peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK fused with substrate peptide,Chaperone protein DnaK fused with substrate peptide, GLYCEROL, ... | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Intermediates in allosteric equilibria of DnaK-ATP interactions with substrate peptides
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7KRW
 
 | |
7KO2
 
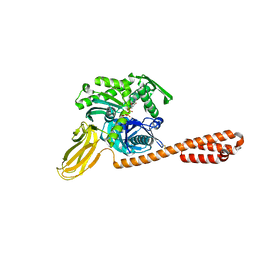 | |
7KRU
 
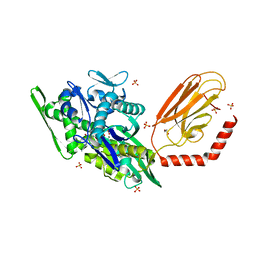 | |
7KRV
 
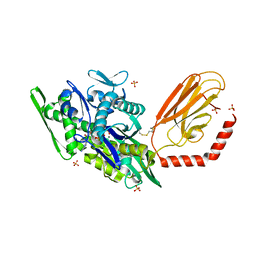 | | Stimulating state of disulfide-bridged Hsp70 DnaK | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK fused with substrate peptide, MAGNESIUM ION, ... | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Conformational equilibria in allosteric control of Hsp70 chaperones.
Mol.Cell, 81, 2021
|
|
7KRT
 
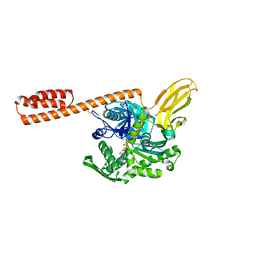 | | Restraining state of a truncated Hsp70 DnaK | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, MAGNESIUM ION | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Conformational equilibria in allosteric control of Hsp70 chaperones.
Mol.Cell, 81, 2021
|
|
7D5I
 
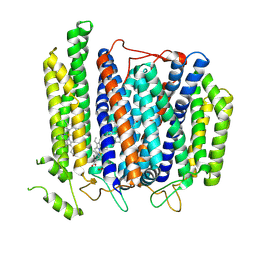 | | Structure of Mycobacterium smegmatis bd complex in the apo-form. | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome D ubiquinol oxidase subunit 1, HEME B/C, ... | | Authors: | Wang, W, Gong, H, Gao, Y, Zhou, X, Rao, Z. | | Deposit date: | 2020-09-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structure of mycobacterial cytochrome bd reveals two oxygen access channels.
Nat Commun, 12, 2021
|
|
8K8T
 
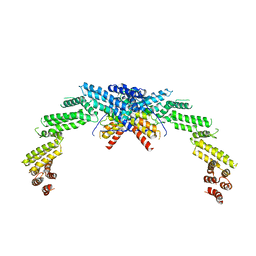 | | Structure of CUL3-RBX1-KLHL22 complex | | Descriptor: | Cullin-3, Kelch-like protein 22 | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
1L7O
 
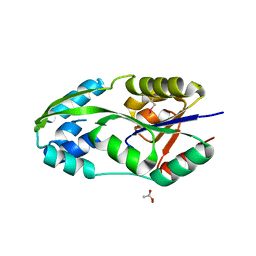 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE IN APO FORM | | Descriptor: | ACETIC ACID, PHOSPHOSERINE PHOSPHATASE, ZINC ION | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7P
 
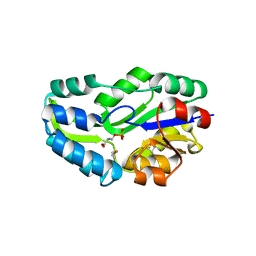 | | SUBSTRATE BOUND PHOSPHOSERINE PHOSPHATASE COMPLEX STRUCTURE | | Descriptor: | PHOSPHATE ION, PHOSPHOSERINE, PHOSPHOSERINE PHOSPHATASE | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7N
 
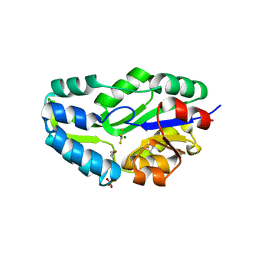 | | TRANSITION STATE ANALOGUE OF PHOSPHOSERINE PHOSPHATASE (ALUMINUM FLUORIDE COMPLEX) | | Descriptor: | ALUMINUM FLUORIDE, MAGNESIUM ION, PHOSPHOSERINE PHOSPHATASE, ... | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1MRZ
 
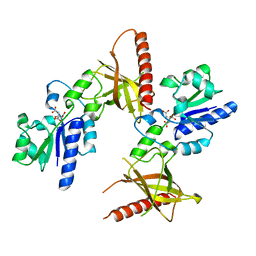 | | Crystal structure of a flavin binding protein from Thermotoga Maritima, TM379 | | Descriptor: | CITRIC ACID, Riboflavin kinase/FMN adenylyltransferase | | Authors: | Wang, W, Kim, R, Jancarik, J, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-09-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a flavin-binding protein from Thermotoga Maritima
Proteins, 52, 2003
|
|
4FXX
 
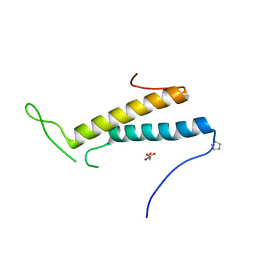 | | Structure of SF1 coiled-coil domain | | Descriptor: | IMIDAZOLE, MALONATE ION, Splicing factor 1 | | Authors: | Gupta, A, Bauer, W.J, Wang, W, Kielkopf, C.L. | | Deposit date: | 2012-07-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4801 Å) | | Cite: | Structure of Phosphorylated SF1 Bound to U2AF(65) in an Essential Splicing Factor Complex.
Structure, 21, 2013
|
|
4O8N
 
 | | Crystal structure of SthAraf62A, a GH62 family alpha-L-arabinofuranosidase from Streptomyces thermoviolaceus, in the apoprotein form | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, CALCIUM ION, ... | | Authors: | Stogios, P.J, Wang, W, Xu, X, Cui, H, Master, E, Savchenko, A. | | Deposit date: | 2013-12-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6476 Å) | | Cite: | Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 alpha-l-arabinofuranosidase from Streptomyces thermoviolaceus.
Appl.Environ.Microbiol., 80, 2014
|
|
4O8P
 
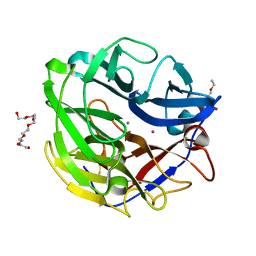 | | Crystal structure of SthAraf62A, a GH62 family alpha-L-arabinofuranosidase from Streptomyces thermoviolaceus, bound to xylotetraose | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Alpha-L-arabinofuranosidase, CALCIUM ION, ... | | Authors: | Stogios, P.J, Wang, W, Xu, X, Cui, H, Master, E, Savchenko, A. | | Deposit date: | 2013-12-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.557 Å) | | Cite: | Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 alpha-l-arabinofuranosidase from Streptomyces thermoviolaceus.
Appl.Environ.Microbiol., 80, 2014
|
|
3SOB
 
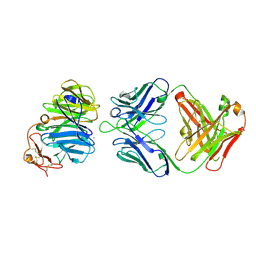 | | The structure of the first YWTD beta propeller domain of LRP6 in complex with a FAB | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 6, antibody heavy chain, ... | | Authors: | Wang, W, Bourhis, E, Tam, C, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
3SOQ
 
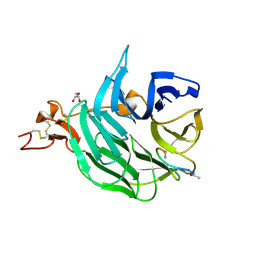 | | The structure of the first YWTD beta propeller domain of LRP6 in complex with a DKK1 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dickkopf-related protein 1, ... | | Authors: | Wang, W, Bourhis, E, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
3SOV
 
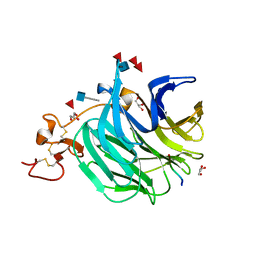 | | The structure of a beta propeller domain in complex with peptide S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Low-density lipoprotein receptor-related protein 6, ... | | Authors: | Wang, W, Bourhis, E, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
4XZC
 
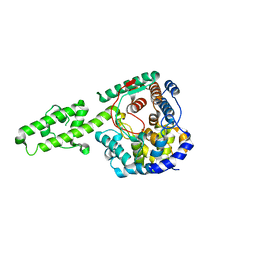 | | The crystal structure of Kupe virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZE
 
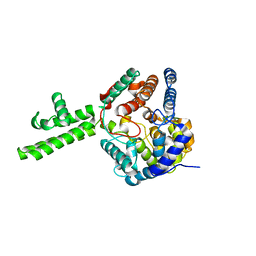 | | The crystal structure of Hazara virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZA
 
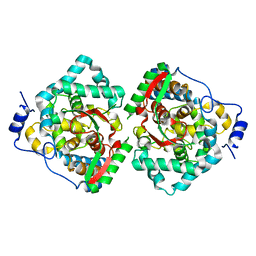 | | The crystal structure of Erve virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZ8
 
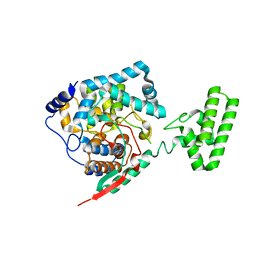 | | The crystal structure of Erve virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
