6AGI
 
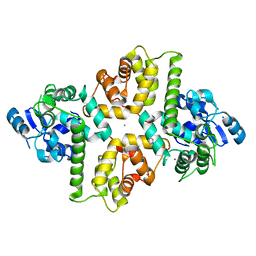 | | Crystal Structure of EFHA2 in Ca-binding State | | Descriptor: | CALCIUM ION, Calcium uptake protein 3, mitochondrial, ... | | Authors: | Yangfei, X, Xue, Y, Yuequan, S. | | Deposit date: | 2018-08-11 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Dimerization of MICU Proteins Controls Ca2+Influx through the Mitochondrial Ca2+Uniporter.
Cell Rep, 26, 2019
|
|
6AGH
 
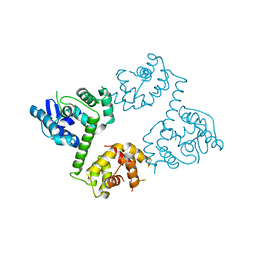 | | Crystal structure of EFHA1 in Apo-State | | Descriptor: | Calcium uptake protein 2, mitochondrial | | Authors: | Yangfei, X, Xue, Y, Yuequan, S. | | Deposit date: | 2018-08-11 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.742 Å) | | Cite: | Dimerization of MICU Proteins Controls Ca2+Influx through the Mitochondrial Ca2+Uniporter.
Cell Rep, 26, 2019
|
|
5KCM
 
 | | Crystal structure of iron-sulfur cluster containing photolyase PhrB mutant I51W | | Descriptor: | (6-4) photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, X, Bowatte, K, Zhang, F, Lamparter, T. | | Deposit date: | 2016-06-06 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
5TUW
 
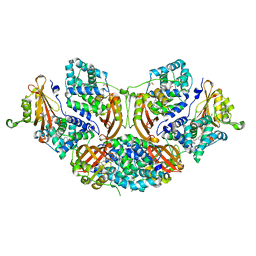 | | Crystal structure of Orange Carotenoid Protein with partial loss of 3'OH Echinenone chromophore | | Descriptor: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, GLYCEROL, Orange carotenoid-binding protein | | Authors: | Yang, X, Bandara, S, Ren, Z. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Photoactivation mechanism of a carotenoid-based photoreceptor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TV0
 
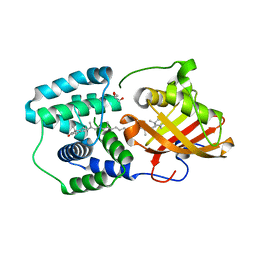 | | crystal structure and light induced structural changes in orange carotenoid protein bound with 3 'OH echinenone | | Descriptor: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, GLYCEROL, Orange carotenoid-binding protein | | Authors: | Yang, X, Bandara, S, Ren, Z. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Photoactivation mechanism of a carotenoid-based photoreceptor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2BTP
 
 | | 14-3-3 Protein Theta (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN TAU, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Elkins, J.M, Johansson, A.C.E, Smee, C, Yang, X, Sundstrom, M, Edwards, A, Arrowsmith, C, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-05 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
6PZT
 
 | | cryo-EM structure of human NKCC1 | | Descriptor: | Solute carrier family 12 member 2 | | Authors: | Cao, E, Wang, Q, Yang, X. | | Deposit date: | 2019-08-01 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of the human cation-chloride cotransporter NKCC1 determined by single-particle electron cryo-microscopy.
Nat Commun, 11, 2020
|
|
2ODB
 
 | | The crystal structure of human cdc42 in complex with the CRIB domain of human p21-activated kinase 6 (PAK6) | | Descriptor: | CHLORIDE ION, Human Cell Division Cycle 42 (CDC42), MAGNESIUM ION, ... | | Authors: | Ugochukwu, E, Yang, X, Elkins, J, Soundararajan, M, Pike, A.C.W, Eswaran, J, Burgess, N, Debreczeni, J.E, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Gileadi, O, von Delft, F, Knapp, S, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-22 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of human cdc42 in complex with the CRIB domain of human p21-activated kinase 6 (PAK6)
To be Published
|
|
2BMJ
 
 | | GTPase like domain of Centaurin Gamma 1 (Human) | | Descriptor: | CENTAURIN GAMMA 1 | | Authors: | Yang, X, Elkins, J.M, Soundararajan, M, Arrowsmith, C, Edwards, A, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-14 | | Release date: | 2005-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Centaurin Gamma-1 Gtpase-Like Domain Functions as an Ntpase.
Biochem.J., 401, 2007
|
|
2C63
 
 | | 14-3-3 Protein Eta (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN ETA, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Elkins, J.M, Yang, X, Smee, C.E.A, Johansson, C, Sundstrom, M, Edwards, A, Weigelt, J, Arrowsmith, C, Doyle, D.A. | | Deposit date: | 2005-11-07 | | Release date: | 2005-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2C23
 
 | | 14-3-3 Protein Beta (Human) in complex with exoenzyme S peptide | | Descriptor: | 14-3-3 BETA/ALPHA, EXOENZYME S PEPTIDE | | Authors: | Elkins, J.M, Schoch, G.A, Yang, X, Sundstrom, M, Arrowsmith, C, Edwards, A, Doyle, D.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2C74
 
 | | 14-3-3 Protein Eta (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN ETA, CITRIC ACID, CONSENSUS PEPTIDE MODE 1 FOR 14-3-3 PROTEINS | | Authors: | Elkins, J.M, Yang, X, Smee, C.E.A, Johansson, C, Sundstrom, M, Edwards, A, Weigelt, J, Arrowsmith, C, Doyle, D.A. | | Deposit date: | 2005-11-17 | | Release date: | 2005-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for protein-protein interactions in the 14-3-3 protein family.
Proc. Natl. Acad. Sci. U.S.A., 103, 2006
|
|
4NAA
 
 | | Crystal structure of UVB photoreceptor UVR8 from Arabidopsis thaliana and UV-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Ren, Z, Zhao, K.H. | | Deposit date: | 2013-10-22 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
4NC4
 
 | | Crystal structure of photoreceptor AtUVR8 mutant W285F and light-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
4NBM
 
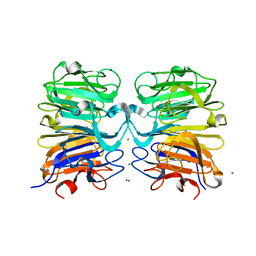 | | Crystal structure of UVB photoreceptor UVR8 and light-induced structural changes at 180K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
5D2N
 
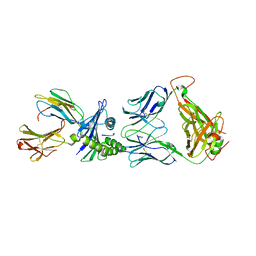 | | Crystal structure of C25-NLV-HLA-A2 complex | | Descriptor: | ASN-LEU-VAL-PRO-MET-VAL-ALA-THR-VAL, Beta-2-microglobulin, C25 alpha, ... | | Authors: | Mariuzza, R.A, Yang, X. | | Deposit date: | 2015-08-05 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural Basis for Clonal Diversity of the Public T Cell Response to a Dominant Human Cytomegalovirus Epitope.
J.Biol.Chem., 290, 2015
|
|
7MCH
 
 | |
9GEK
 
 | | Structure of the FAST1-FAST2-RAP module from human FASTKD4 by carrier-driven crystallisation with maltose binding protein from E. coli. | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Maltose/maltodextrin-binding periplasmic protein,FAST kinase domain-containing protein 4,FAST kinase domain-containing protein 4, PHOSPHATE ION, ... | | Authors: | Hothorn, M, Lau, K, Yang, X. | | Deposit date: | 2024-08-07 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Vsr-like protein FASTKD4 regulates the stability and polyadenylation of the MT-ND3 mRNA.
Nucleic Acids Res., 53, 2025
|
|
7MC4
 
 | |
7C2N
 
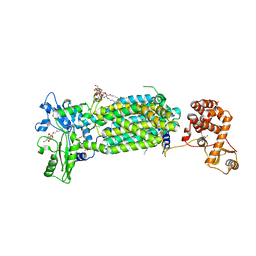 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SPIRO | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1'-(2,3-dihydro-1,4-benzodioxin-6-ylmethyl)spiro[6,7-dihydrothieno[3,2-c]pyran-4,4'-piperidine], Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7C2M
 
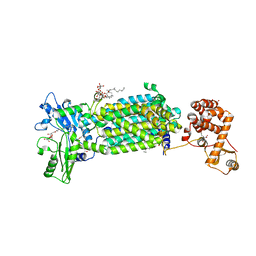 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with NITD-349 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Chimera of drug exporters of the RND superfamily-like protein and Endolysin, N-(4,4-dimethylcyclohexyl)-4,6-bis(fluoranyl)-1H-indole-2-carboxamide, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7XRW
 
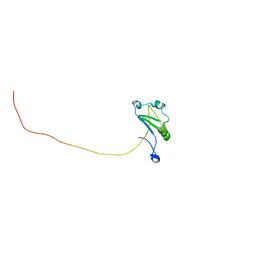 | | Solution structure of T. brucei RAP1 | | Descriptor: | Repressor activator protein 1 | | Authors: | Yang, X, Pan, X.H, Ji, Z.Y, Wong, K.B, Zhang, M.J, Zhao, Y.X. | | Deposit date: | 2022-05-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The RRM-mediated RNA binding activity in T. brucei RAP1 is essential for VSG monoallelic expression.
Nat Commun, 14, 2023
|
|
8F71
 
 | |
5XGH
 
 | | Crystal structure of PI3K complex with an inhibitor | | Descriptor: | 3-[(4-fluorophenyl)methylamino]-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Song, K, Yang, X, Zhao, Y, Jian, Z. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | New Insights into PI3K Inhibitor Design using X-ray Structures of PI3K alpha Complexed with a Potent Lead Compound.
Sci Rep, 7, 2017
|
|
5TUX
 
 | | crystal structure and light induced structural changes in orange carotenoid protein bound with echinenone | | Descriptor: | GLYCEROL, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | Authors: | Yang, X, Bandara, S, Ren, Z. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Photoactivation mechanism of a carotenoid-based photoreceptor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
