3ZD4
 
 | |
2G04
 
 | | Crystal structure of fatty acid-CoA racemase from Mycobacterium tuberculosis H37Rv | | Descriptor: | PROBABLE FATTY-ACID-CoA RACEMASE FAR | | Authors: | Lee, K.S, Park, S.M, Rhee, K.H, Bang, W.G, Hwang, K.Y, Chi, Y.M. | | Deposit date: | 2006-02-11 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of fatty acid-CoA racemase from Mycobacterium tuberculosis H37Rv
Proteins, 64, 2006
|
|
2ISZ
 
 | | Crystal structure of a two-domain IdeR-DNA complex crystal form I | | Descriptor: | Iron-dependent repressor ideR, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
2IT0
 
 | | Crystal structure of a two-domain IdeR-DNA complex crystal form II | | Descriptor: | ACETATE ION, Iron-dependent repressor ideR, NICKEL (II) ION, ... | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
2ISY
 
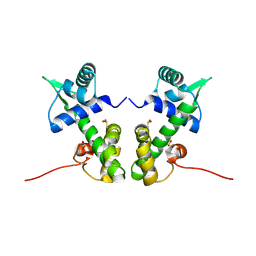 | | Crystal structure of the nickel-activated two-domain iron-dependent regulator (IdeR) | | Descriptor: | Iron-dependent repressor ideR, NICKEL (II) ION, PHOSPHATE ION | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
3TEC
 
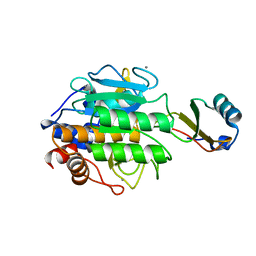 | | CALCIUM BINDING TO THERMITASE. CRYSTALLOGRAPHIC STUDIES OF THERMITASE AT 0, 5 AND 100 MM CALCIUM | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Kalk, K.H, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calcium binding to thermitase. Crystallographic studies of thermitase at 0, 5, and 100 mM calcium.
J.Biol.Chem., 266, 1991
|
|
2MAD
 
 | |
2VRT
 
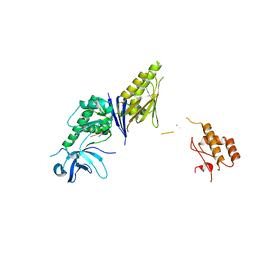 | | Crystal Structure of E. coli RNase E possessing M1 RNA fragments - Catalytic Domain | | Descriptor: | 5'-R(*UP*UP)-3', 5'-R(*UP*UP*GP)-3', RIBONUCLEASE E, ... | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Garman, E.F, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-04-14 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
2VMK
 
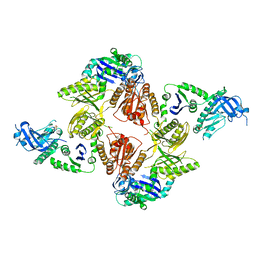 | | Crystal Structure of E. coli RNase E Apoprotein - Catalytic Domain | | Descriptor: | RIBONUCLEASE E, SULFATE ION, ZINC ION | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
2SBT
 
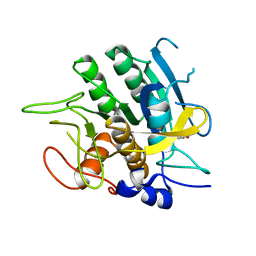 | | A COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF SUBTILISIN BPN AND SUBTILISIN NOVO | | Descriptor: | ACETONE, SUBTILISIN NOVO | | Authors: | Drenth, J, Hol, W.G.J, Jansonius, J.N, Koekoek, R. | | Deposit date: | 1976-09-07 | | Release date: | 1976-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A comparison of the three-dimensional structures of subtilisin BPN' and subtilisin novo.
Cold Spring Harbor Symp.Quant.Biol., 36, 1972
|
|
1BI0
 
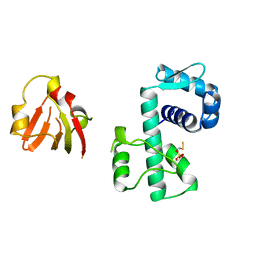 | | STRUCTURE OF APO-AND HOLO-DIPHTHERIA TOXIN REPRESSOR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR, SULFATE ION, ZINC ION | | Authors: | Pohl, E, Hol, W.G. | | Deposit date: | 1998-06-21 | | Release date: | 1999-07-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR.
J.Biol.Chem., 273, 1998
|
|
2X0N
 
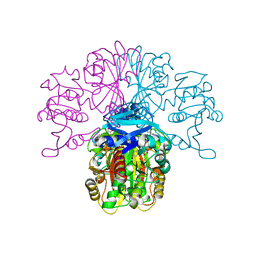 | | Structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei determined from Laue data | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, GLYCOSOMAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Vellieux, F.M.D, Hajdu, J, Hol, W.G.J. | | Deposit date: | 2009-12-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma Brucei Determined from Laue Data.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
6HG0
 
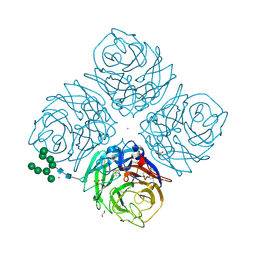 | | Influenza A Virus N9 Neuraminidase complex with NANA (Tern/Australia). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
6IIG
 
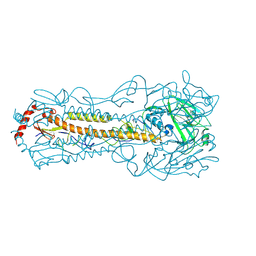 | |
6HCX
 
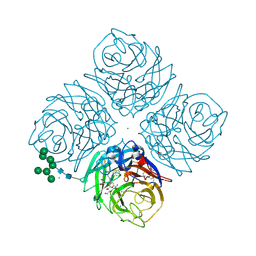 | | Influenza Virus N9 Neuraminidase A complex with Zanamivir molecule (Tern). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
6HGB
 
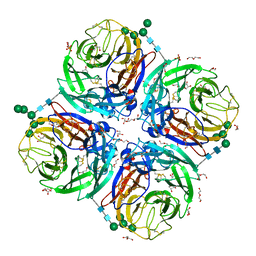 | | Influenza A virus N6 neuraminidase native structure (Duck/England/56). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-23 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
6HEB
 
 | | Influenza A Virus N9 Neuraminidase complex with Oseltamivir (Tern). | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-20 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site.
To Be Published
|
|
6HFC
 
 | | Influenza A Virus N9 Neuraminidase Native (Tern). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
6HFY
 
 | | Influenza A virus N6 neuraminidase complex with DANA (Duck/England/56). | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
6HG5
 
 | | Influenza A virus N6 neuraminidase complex with Oseltamivir (Duck/England/56). | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site.
To Be Published
|
|
6KCJ
 
 | |
1EAF
 
 | |
1EAA
 
 | |
1EAE
 
 | |
1EFI
 
 | |
