6IP8
 
 | | Cryo-EM structure of the HCV IRES dependently initiated CMV-stalled 80S ribosome (Structure iv) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6IP6
 
 | | Cryo-EM structure of the CMV-stalled human 80S ribosome with HCV IRES (Structure iii) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6IP5
 
 | | Cryo-EM structure of the CMV-stalled human 80S ribosome (Structure ii) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6JLZ
 
 | | P-eIF2a - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, PHOSPHATE ION, Probable translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-01 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
6JLY
 
 | | eIF2a - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, PHOSPHATE ION, Probable translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
5AZD
 
 | | Crystal structure of thermophilic rhodopsin. | | Descriptor: | Bacteriorhodopsin | | Authors: | Mizutani, K, Hashimoto, N, Tsukamoto, T, Yamashita, K, Yamamoto, M, Sudo, Y, Murata, T. | | Deposit date: | 2015-09-30 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic structure of thermophilic rhodopsin: implications for high thermal stability and optogenetic availability.
To Be Published
|
|
6K72
 
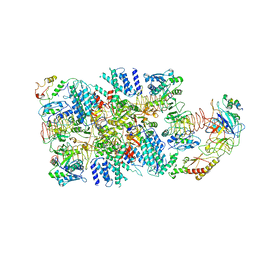 | | eIF2(aP) - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Kashiwagi, K, Yokoyama, T, Ito, T. | | Deposit date: | 2019-06-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
6K71
 
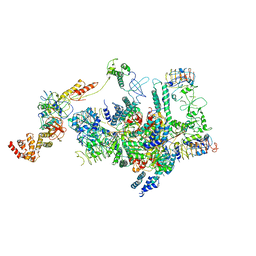 | | eIF2 - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Kashiwagi, K, Yokoyama, T, Ito, T. | | Deposit date: | 2019-06-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
5B04
 
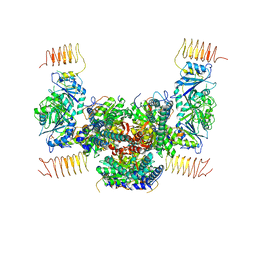 | | Crystal structure of the eukaryotic translation initiation factor 2B from Schizosaccharomyces pombe | | Descriptor: | PHOSPHATE ION, Probable translation initiation factor eIF-2B subunit beta, Probable translation initiation factor eIF-2B subunit delta, ... | | Authors: | Kashiwagi, K, Ito, T, Yokoyama, S. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Crystal structure of eukaryotic translation initiation factor 2B
Nature, 531, 2016
|
|
5TR6
 
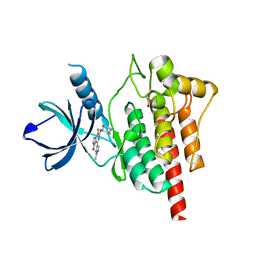 | | Discovery of TAK-659, an Orally Available Investigational Inhibitor of Spleen Tyrosine Kinase (SYK) | | Descriptor: | 1,2-ETHANEDIOL, 6-{[(1R,2S)-2-aminocyclohexyl]amino}-7-fluoro-4-(1-methyl-1H-pyrazol-4-yl)-1,2-dihydro-3H-pyrrolo[3,4-c]pyridin-3-one, Tyrosine-protein kinase SYK | | Authors: | Yano, J, Jennings, A, Lam, B, Hoffman, I.D. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of TAK-659 an orally available investigational inhibitor of Spleen Tyrosine Kinase (SYK).
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5TT7
 
 | | Discovery of TAK-659, an Orally Available Investigational Inhibitor of Spleen Tyrosine Kinase (SYK) | | Descriptor: | 2-{[(1R,2S)-2-aminocyclohexyl]amino}-4-[(3-methylphenyl)amino]-6,7-dihydro-5H-pyrrolo[3,4-d]pyrimidin-5-one, Tyrosine-protein kinase SYK | | Authors: | Yano, J, Jennings, A, Lam, B, Hoffman, I.D. | | Deposit date: | 2016-11-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of TAK-659 an orally available investigational inhibitor of Spleen Tyrosine Kinase (SYK).
Bioorg. Med. Chem. Lett., 26, 2016
|
|
7F5K
 
 | | Crystal structure of TCR4-1 ectodomain | | Descriptor: | T cell receptor alpha chain, T cell receptor beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.0000298 Å) | | Cite: | Symbiotic bacteria-dependent expansion of MR1-reactive T cells causes autoimmunity in the absence of Bcl11b.
Nat Commun, 13, 2022
|
|
7F66
 
 | | eIF2B-SFSV NSs-1-eIF2 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, Non-structural protein NS-S, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7F67
 
 | | eIF2B-SFSV NSs-2-eIF2 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, Non-structural protein NS-S, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7FBV
 
 | |
7F64
 
 | | eIF2B-SFSV NSs | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7FBR
 
 | |
7VLK
 
 | | eIF2B-SFSV NSs C2-imposed | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-10-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
2Z0Z
 
 | | Crystal structure of putative acetyltransferase | | Descriptor: | Putative uncharacterized protein TTHA1799, SULFATE ION | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genetic Encoding of 3-Iodo-l-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography
Structure, 17, 2009
|
|
2DDT
 
 | | Crystal structure of sphingomyelinase from Bacillus cereus with magnesium ion | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ago, H, Oda, M, Tsuge, H, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-02 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the sphingomyelin phosphodiesterase activity in neutral sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
2Z10
 
 | | Crystal structure of putative acetyltransferase | | Descriptor: | Ribosomal-protein-alanine acetyltransferase | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Genetic Encoding of 3-Iodo-l-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography
Structure, 17, 2009
|
|
2ZXV
 
 | | Crystal structure of putative acetyltransferase from T. thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHA1799 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genetic Encoding of 3-Iodo-l-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography
Structure, 17, 2009
|
|
2DK4
 
 | | Solution structure of Splicing Factor Motif in Pre-mRNA splicing factor 18 (hPRP18) | | Descriptor: | Pre-mRNA-splicing factor 18 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the splicing factor motif of the human Prp18 protein.
Proteins, 80, 2012
|
|
2E61
 
 | | Solution structure of the zf-CW domain in zinc finger CW-type PWWP domain protein 1 | | Descriptor: | ZINC ION, Zinc finger CW-type PWWP domain protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-25 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the zinc finger CW domain as a histone modification reader
Structure, 18, 2010
|
|
7X7M
 
 | | Lumazine Synthase from Aquifex aeolicus | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Sobhy, M.A, Hamdan, S.M. | | Deposit date: | 2022-03-09 | | Release date: | 2022-10-12 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Cryo-electron structures of the extreme thermostable enzymes Sulfur Oxygenase Reductase and Lumazine Synthase.
Plos One, 17, 2022
|
|
