4AZR
 
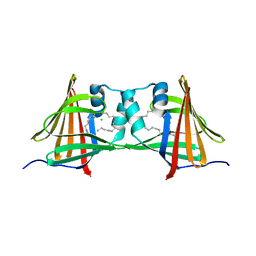 | | Human epidermal fatty acid-binding protein (FABP5) in complex with the endocannabinoid anandamide | | Descriptor: | CHLORIDE ION, FATTY ACID-BINDING PROTEIN, EPIDERMAL, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZN
 
 | | Murine epidermal fatty acid-binding protein (FABP5), apo form, poly- his tag-mediated crystal packing | | Descriptor: | FATTY ACID-BINDING PROTEIN, EPIDERMAL | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZM
 
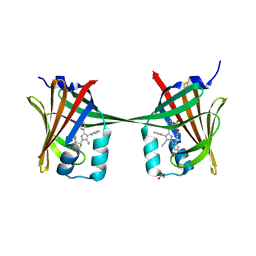 | | Human epidermal fatty acid-binding protein (FABP5) in complex with the inhibitor BMS-309413 | | Descriptor: | ((2'-(5-ETHYL-3,4-DIPHENYL-1H-PYRAZOL-1-YL)-3-BIPHENYLYL)OXY)ACETIC ACID, FATTY ACID-BINDING PROTEIN, EPIDERMAL, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZO
 
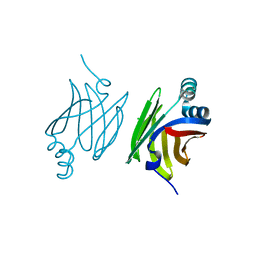 | | Murine epidermal fatty acid-binding protein (FABP5), apo form, poly- his tag removed | | Descriptor: | CHLORIDE ION, FATTY ACID-BINDING PROTEIN, EPIDERMAL | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ZUZ
 
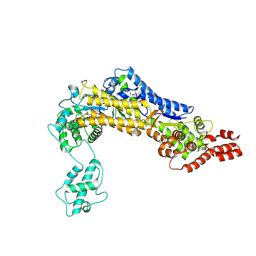 | | SidC 1-871 | | Descriptor: | SidC | | Authors: | Luo, X, Wasilko, D.J, Liu, Y, Sun, J, Wu, X, Luo, Z.-Q, Mao, Y. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of the Legionella Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome.
Plos Pathog., 11, 2015
|
|
1Q6B
 
 | | Solution Structure of the C-terminal Domain of Thermosynechococcus elongatus KaiA (ThKaiA180C); Ensemble of 25 Structures | | Descriptor: | Circadian clock protein KaiA homolog | | Authors: | Vakonakis, I, Sun, J, Golden, S.S, Holzenburg, A, LiWang, A.C. | | Deposit date: | 2003-08-13 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: implications for KaiA-KaiC interaction
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q6A
 
 | | Solution Structure of the C-terminal Domain of Thermosynechococcus elongatus KaiA (ThKaiA180C); Averaged Minimized Structure | | Descriptor: | Circadian clock protein KaiA homolog | | Authors: | Vakonakis, I, Sun, J, Holzenburg, A, Golden, S.S, LiWang, A.C. | | Deposit date: | 2003-08-13 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: Implications for KaiA-KaiC interaction
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5B5X
 
 | | Crystal structure of limiting CO2-inducible protein LCIC | | Descriptor: | SULFATE ION, ZINC ION, limiting CO2-inducible protein LCIC | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Cajar, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.511 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5B60
 
 | | Crystal structure of PtLCIB4 S47R mutant, a homolog of the limiting CO2-inducible protein LCIB | | Descriptor: | CHLORIDE ION, PtLCIB4 S47R mutant, ZINC ION | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Caja, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5B5Y
 
 | | Crystal structure of PtLCIB4, a homolog of the limiting CO2-inducible protein LCIB | | Descriptor: | ACETATE ION, PtLCIB4, ZINC ION | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Cajar, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5B5Z
 
 | | Crystal structure of PtLCIB4 H88A mutant, a homolog of the limiting CO2-inducible protein LCIB | | Descriptor: | PtLCIB4 H88A mutant, ZINC ION | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Caja, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6A67
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody FLD21.140 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLD21.140 Heavy Chain, FLD21.140 Light Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Complementary recognition of the receptor-binding site of highly pathogenic H5N1 influenza viruses by two human neutralizing antibodies.
J. Biol. Chem., 293, 2018
|
|
4Q2C
 
 | | Crystal structure of CRISPR-associated protein | | Descriptor: | CRISPR-associated helicase Cas3, NICKEL (II) ION | | Authors: | Gong, B, Shin, M, Sun, J, van der Oost, J, Kim, J.-S. | | Deposit date: | 2014-04-07 | | Release date: | 2014-11-19 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q2D
 
 | | Crystal Structure of CRISPR-Associated protein in complex with 2'-Deoxyadenosine 5'-Triphosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CRISPR-associated helicase Cas3, MAGNESIUM ION, ... | | Authors: | Gong, B, Shin, M, Sun, J, van der Oost, J, Kim, J.-S. | | Deposit date: | 2014-04-07 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.771 Å) | | Cite: | Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8FO2
 
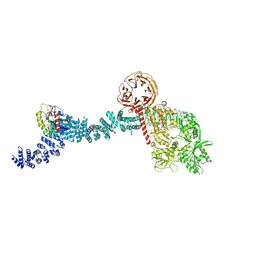 | |
8FO7
 
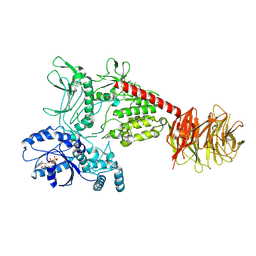 | | Cryo-EM structure of LRRK2 bound to type I inhibitor LRRK2-IN-1 | | Descriptor: | 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM.
Cell Discov, 10, 2024
|
|
8FH3
 
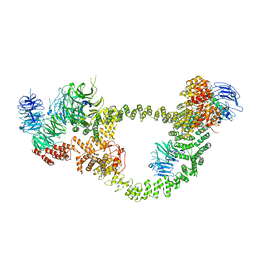 | | Human IFT-A complex structures provide molecular insights into ciliary transport | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Tubby-related protein 3, ... | | Authors: | Jiang, M, Palicharla, V.R, Miller, D, Hwang, S.H, Zhu, H, Hixson, P, Mukhopadhyay, S, Sun, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Human IFT-A complex structures provide molecular insights into ciliary transport.
Cell Res., 33, 2023
|
|
8FGW
 
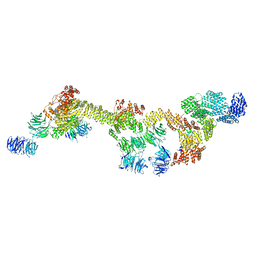 | | Human IFT-A complex structures provide molecular insights into ciliary transport | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Intraflagellar transport protein 43 homolog, ... | | Authors: | Jiang, M, Palicharla, V.R, Miller, D, Hwang, S.H, Zhu, H, Hixson, P, Mukhopadhyay, S, Sun, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Human IFT-A complex structures provide molecular insights into ciliary transport.
Cell Res., 33, 2023
|
|
3U9G
 
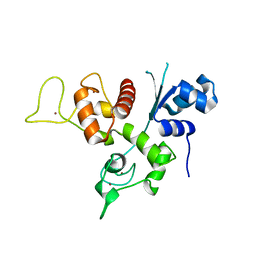 | | Crystal structure of the Zinc finger antiviral protein | | Descriptor: | ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Chen, S, Xu, Y, Zhang, K, Wang, X, Sun, J, Gao, G, Liu, Y. | | Deposit date: | 2011-10-18 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UFB
 
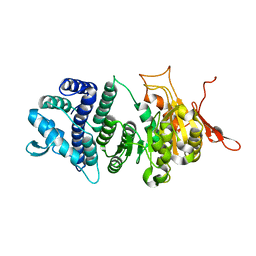 | | Crystal structure of a modification subunit of a putative type I restriction enzyme from Vibrio vulnificus YJ016 | | Descriptor: | Type I restriction-modification system methyltransferase subunit | | Authors: | Park, S.Y, Lee, H.J, Sun, J, Nishi, K, Song, J.M, Kim, J.S. | | Deposit date: | 2011-11-01 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of a modification subunit of a putative type I restriction enzyme from Vibrio vulnificus YJ016
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4GKF
 
 | | Crystal structure and characterization of Cmr5 protein from Pyrococcus furiosus | | Descriptor: | CRISPR system Cmr subunit Cmr5 | | Authors: | Park, J, Sun, J, Park, S, Hwang, H, Park, M, Shin, M.S. | | Deposit date: | 2012-08-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Cmr5 from Pyrococcus furiosus and its functional implications
Febs Lett., 587, 2013
|
|
3OX4
 
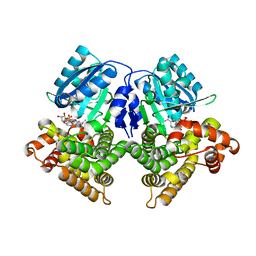 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 complexed with NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
3OWO
 
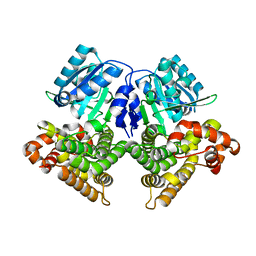 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
3PST
 
 | |
3PSP
 
 | |
