3ALU
 
 | | Crystal structure of CEL-IV complexed with Raffinose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Lectin CEL-IV, ... | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
3ALT
 
 | | Crystal structure of CEL-IV complexed with Melibiose | | Descriptor: | CALCIUM ION, Lectin CEL-IV, C-type, ... | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
3AXH
 
 | | Crystal structure of isomaltase in complex with isomaltose | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase IMA1, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2011-04-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae
J.Biosci.Bioeng., 112, 2011
|
|
3AXI
 
 | | Crystal structure of isomaltase in complex with maltose | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase IMA1, alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2011-04-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae
J.Biosci.Bioeng., 112, 2011
|
|
2ZRW
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with FMN and IPP. | | Descriptor: | FLAVIN MONONUCLEOTIDE, ISOPENTYL PYROPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, ... | | Authors: | Unno, H, Yamashita, S, Ikeda, Y, Sekiguchi, S, Yoshida, N, Yoshimura, T, Kusunoki, M, Nakayama, T, Nishino, T, Hemmi, H. | | Deposit date: | 2008-09-01 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
J.Biol.Chem., 284, 2009
|
|
2ZOG
 
 | | Crystal structure of mouse carnosinase CN2 complexed with ZN and bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Cytosolic non-specific dipeptidase, ZINC ION | | Authors: | Unno, H, Yamashita, T, Okumura, N, Kusunoki, M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for substrate recognition and hydrolysis by mouse carnosinase CN2.
J.Biol.Chem., 283, 2008
|
|
2ZRU
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Isopentenyl-diphosphate delta-isomerase | | Authors: | Unno, H, Yamashita, S, Ikeda, Y, Sekiguchi, S, Yoshida, N, Yoshimura, T, Kusunoki, M, Nakayama, T, Nishino, T, Hemmi, H. | | Deposit date: | 2008-09-01 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
J.Biol.Chem., 284, 2009
|
|
3A4A
 
 | | Crystal structure of isomaltase from Saccharomyces cerevisiae | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase, alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose
Febs J., 277, 2010
|
|
1OX1
 
 | | crystal structure of the bovine trypsin complex with a synthetic 11 peptide inhibitor | | Descriptor: | 11-mer peptide, CALCIUM ION, Trypsinogen, ... | | Authors: | Wu, G, Huang, Y, Zhu, G, Huang, Q, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-03-31 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of the bovine trypsin complex with a synthetic 11 peptide inhibitor
To be published
|
|
1OMY
 
 | | Crystal Structure of a Recombinant alpha-insect Toxin BmKaIT1 from the scorpion Buthus martensii Karsch | | Descriptor: | ACETIC ACID, Alpha-neurotoxin TX12, CHLORIDE ION | | Authors: | Huang, Y, Huang, Q, Chen, H, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary crystallographic study of rBmKalphaIT1, a recombinant alpha-insect toxin from the scorpion Buthus martensii Karsch.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1ITC
 
 | | Beta-Amylase from Bacillus cereus var. mycoides Complexed with Maltopentaose | | Descriptor: | ACETIC ACID, Beta-Amylase, CALCIUM ION, ... | | Authors: | Miyake, H, Kurisu, G, Kusunoki, M, Nishimura, S, Kitamura, S, Nitta, Y. | | Deposit date: | 2002-01-17 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Catalytic Site Mutant of beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltopentaose
BIOCHEMISTRY, 42, 2003
|
|
1J18
 
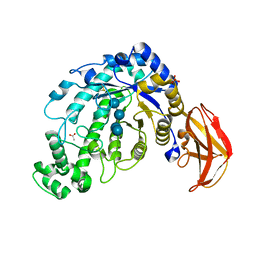 | | Crystal Structure of a Beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltose | | Descriptor: | ACETIC ACID, Beta-amylase, CALCIUM ION, ... | | Authors: | Miyake, H, Kurisu, G, Kusunoki, M, Nishimura, S, Kitamura, S, Nitta, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Catalytic Site Mutant of beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltopentaose
BIOCHEMISTRY, 42, 2003
|
|
3TKK
 
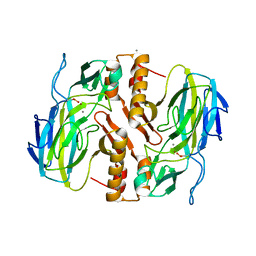 | | Crystal Structure Analysis of a recombinant predicted acetamidase/ formamidase from the thermophile thermoanaerobacter tengcongensis | | Descriptor: | CALCIUM ION, Predicted acetamidase/formamidase, ZINC ION | | Authors: | Qian, M, Huang, Q, Wu, G, Lai, L, Tang, Y, Pei, J, Kusunoki, M. | | Deposit date: | 2011-08-26 | | Release date: | 2011-11-16 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure Analysis of a Recombinant Predicted Acetamidase/Formamidase from the Thermophile Thermoanaerobacter tengcongensis.
PROTEIN J., 31, 2012
|
|
3WJ2
 
 | | Crystal structure of ESTFA (FE-lacking apo form) | | Descriptor: | Carboxylesterase | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
3WMH
 
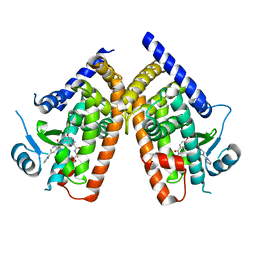 | |
3WJ1
 
 | | Crystal structure of SSHESTI | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
1GEE
 
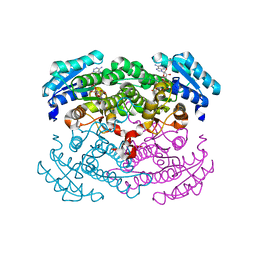 | | Crystal structure of glucose dehydrogenase mutant Q252L complexed with NAD+ | | Descriptor: | GLUCOSE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-11-07 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of stability-increasing mutants of glucose dehydrogenase
To be Published
|
|
1G6K
 
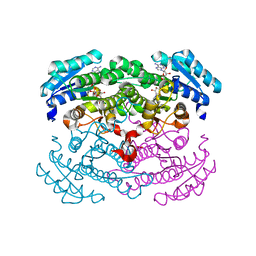 | | Crystal structure of glucose dehydrogenase mutant E96A complexed with NAD+ | | Descriptor: | GLUCOSE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-11-06 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of stability-increasing mutants of glucose dehydrogenase
To be Published
|
|
1GEG
 
 | | CRYATAL STRUCTURE ANALYSIS OF MESO-2,3-BUTANEDIOL DEHYDROGENASE | | Descriptor: | ACETOIN REDUCTASE, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Otagiri, M, Kurisu, G, Ui, S, Kusunoki, M. | | Deposit date: | 2000-11-10 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of meso-2,3-butanediol dehydrogenase in a complex with NAD+ and inhibitor mercaptoethanol at 1.7 A resolution for understanding of chiral substrate recognition mechanisms.
J.Biochem., 129, 2001
|
|
1GCO
 
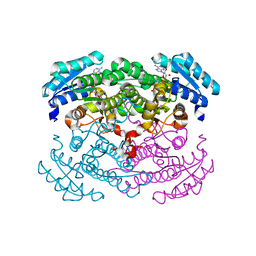 | | CRYSTAL STRUCTURE OF GLUCOSE DEHYDROGENASE COMPLEXED WITH NAD+ | | Descriptor: | GLUCOSE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-08-07 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glucose dehydrogenase from Bacillus megaterium IWG3 at 1.7 A resolution.
J.Biochem., 129, 2001
|
|
1GAQ
 
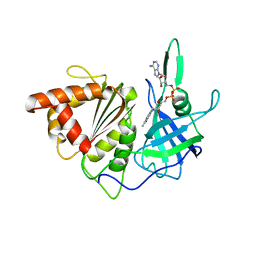 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN FERREDOXIN AND FERREDOXIN-NADP+ REDUCTASE | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I, FERREDOXIN-NADP+ REDUCTASE, ... | | Authors: | Kurisu, G, Kusunoki, M, Hase, T. | | Deposit date: | 2000-05-08 | | Release date: | 2001-02-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of the electron transfer complex between ferredoxin and ferredoxin-NADP(+) reductase.
Nat.Struct.Biol., 8, 2001
|
|
1GAW
 
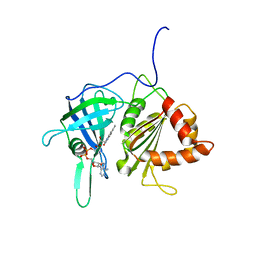 | |
7T71
 
 | | Crystal Structure of Mevalonate 3,5-Bisphosphate Decarboxylase from Picrophilus Torridus | | Descriptor: | Mevalonate 3,5-bisphosphate decarboxylase, OLEIC ACID | | Authors: | Vinokur, J.M, Sawaya, M.R, Cascio, D, Collazo, M, Bowie, J.U. | | Deposit date: | 2021-12-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of mevalonate 3,5-bisphosphate decarboxylase reveals insight into the evolution of decarboxylases in the mevalonate metabolic pathways.
J.Biol.Chem., 298, 2022
|
|
5Y6Q
 
 | |
5ZU2
 
 | | Effect of mutation (R554A) on FAD modification in Aspergillus oryzae RIB40formate oxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mikami, B, Uchida, H, Doubayashi, D. | | Deposit date: | 2018-05-06 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | The microenvironment surrounding FAD mediates its conversion to 8-formyl-FAD in Aspergillus oryzae RIB40 formate oxidase.
J.Biochem., 166, 2019
|
|
