2AEB
 
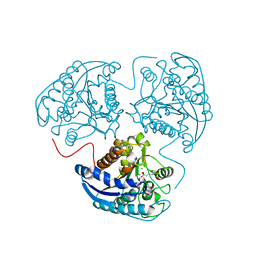 | | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in immune response. | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase 1, MANGANESE (II) ION | | Authors: | Di Costanzo, L, Sabio, G, Mora, A, Rodriguez, P.C, Ochoa, A.C, Centeno, F, Christianson, D.W. | | Deposit date: | 2005-07-21 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in the immune response.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1IVW
 
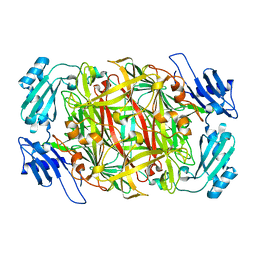 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Late intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVX
 
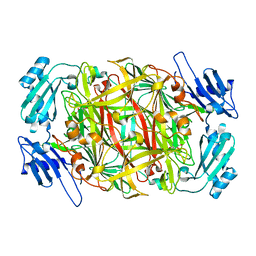 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Holo form generated by biogenesis in crystal. | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVU
 
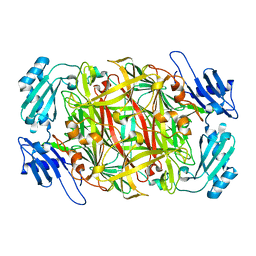 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Initial intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVV
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Early intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1K1V
 
 | | Solution Structure of the DNA-Binding Domain of MafG | | Descriptor: | MafG | | Authors: | Kusunoki, H, Motohashi, H, Katsuoka, F, Morohashi, A, Yamamoto, M, Tanaka, T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of MafG.
Nat.Struct.Biol., 9, 2002
|
|
4A5S
 
 | | CRYSTAL STRUCTURE OF HUMAN DPP4 IN COMPLEX WITH A NOVAL HETEROCYCLIC DPP4 INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[(3S)-3-AMINOPIPERIDIN-1-YL]-5-BENZYL-4-OXO-3-(QUINOLIN-4-YLMETHYL)-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDINE-7-CARBONITRILE, DIPEPTIDYL PEPTIDASE 4 SOLUBLE FORM, ... | | Authors: | Ostermann, N, Kroemer, M, Zink, F, Gerhartz, B, Sutton, J.M, Clark, D.E, Dunsdon, S.J, Fenton, G, Fillmore, A, Harris, N.V, Higgs, C, Hurley, C.A, Krintel, S.L, MacKenzie, R.E, Duttaroy, A, Gangl, E, Maniara, W, Sedrani, R, Namoto, K, Sirockin, F, Trappe, J, Hassiepen, U, Baeschlin, D.K. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Novel Heterocyclic Dpp-4 Inhibitors for the Treatment of Type 2 Diabetes.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AFL
 
 | | The crystal structure of the ING4 dimerization domain reveals the functional organization of the ING family of chromatin binding proteins. | | Descriptor: | INHIBITOR OF GROWTH PROTEIN 4 | | Authors: | Culurgioni, S, Munoz, I.G, Moreno, A, Palacios, A, Villate, M, Palmero, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-22 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystal Structure of Inhibitor of Growth 4 (Ing4) Dimerization Domain Reveals Functional Organization of Ing Family of Chromatin-Binding Proteins.
J.Biol.Chem., 287, 2012
|
|
4B8S
 
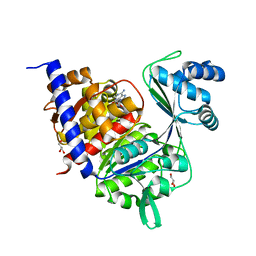 | | Crystal Structure of Thermococcus litoralis ADP-dependent Glucokinase (GK) | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-DEPENDENT GLUCOKINASE, GLYCEROL, ... | | Authors: | Herrera-Morande, A, Rivas-Pardo, J.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal Structure, Saxs and Kinetic Mechanism of Hyperthermophilic Adp-Dependent Glucokinase from Thermococcus Litoralis Reveal a Conserved Mechanism for Catalysis.
Plos One, 8, 2013
|
|
3W6H
 
 | | Crystal structure of 19F probe-labeled hCAI in complex with acetazolamide | | Descriptor: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 1, ... | | Authors: | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.964 Å) | | Cite: | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
3W6I
 
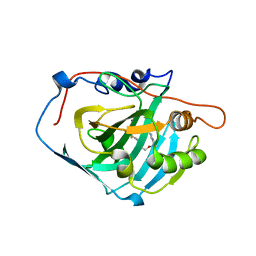 | | Crystal structure of 19F probe-labeled hCAI | | Descriptor: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, Carbonic anhydrase 1, ZINC ION | | Authors: | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
5O5X
 
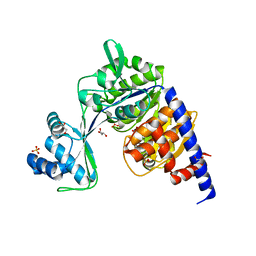 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | Descriptor: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, SULFATE ION | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5O5Y
 
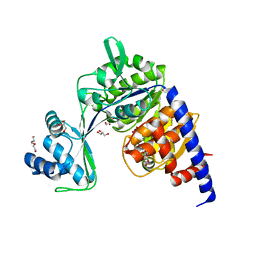 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | Descriptor: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.915 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
4AXP
 
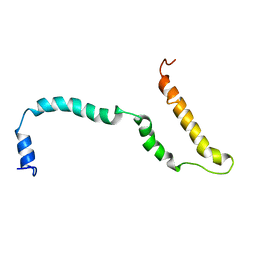 | | NMR structure of Hsp12, a protein induced by and required for dietary restriction-induced lifespan extension in yeast. | | Descriptor: | 12 KDA HEAT SHOCK PROTEIN | | Authors: | Herbert, A.P, Riesen, M, Bloxam, L, Kosmidou, E, Wareing, B.M, Johnson, J.R, Phelan, M.M, Pennington, S.R, Lian, L.Y, Morgan, A. | | Deposit date: | 2012-06-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Hsp12, a Protein Induced by and Required for Dietary Restriction-Induced Lifespan Extension in Yeast.
Plos One, 7, 2012
|
|
5NWJ
 
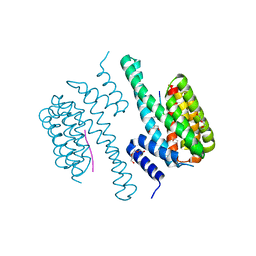 | | 14-3-3c in complex with CPP7 | | Descriptor: | 14-3-3-like protein C, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saponaro, A, Porro, A, Chaves-Sanjuan, A, Nardini, M, Thiel, G, Moroni, A. | | Deposit date: | 2017-05-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Fusicoccin Activates KAT1 Channels by Stabilizing Their Interaction with 14-3-3 Proteins.
Plant Cell, 29, 2017
|
|
8SV9
 
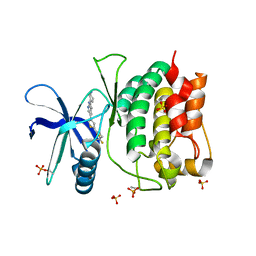 | | Crystal structure of ULK1 kinase domain with inhibitor MR-2088 | | Descriptor: | (4P)-4-[(2P)-2-(1,2,5,6-tetrahydropyridin-3-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N-(2,2,2-trifluoroethyl)thiophene-2-carboxamide, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-05-15 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of potent and selective ULK1/2 inhibitors based on 7-azaindole scaffold with favorable in vivo properties.
Eur.J.Med.Chem., 266, 2024
|
|
2M1R
 
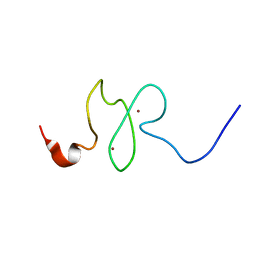 | | PHD domain of ING4 N214D mutant | | Descriptor: | Inhibitor of growth protein 4, ZINC ION | | Authors: | Blanco, F.J. | | Deposit date: | 2012-12-05 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Functional impact of cancer-associated mutations in the tumor suppressor protein ING4.
Carcinogenesis, 31, 2010
|
|
4X68
 
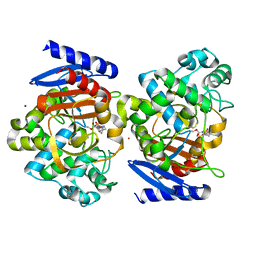 | | Crystal Structure of OP0595 complexed with AmpC | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, NICKEL (II) ION | | Authors: | Yamada, M, Watanabe, T. | | Deposit date: | 2014-12-07 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam 'enhancer'
J.Antimicrob.Chemother., 70, 2015
|
|
4X69
 
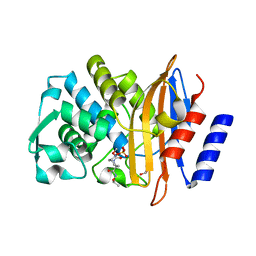 | | Crystal structure of OP0595 complexed with CTX-M-44 | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase Toho-1 | | Authors: | Yamada, M, Watanabe, T. | | Deposit date: | 2014-12-07 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam 'enhancer'
J.Antimicrob.Chemother., 70, 2015
|
|
6Q3S
 
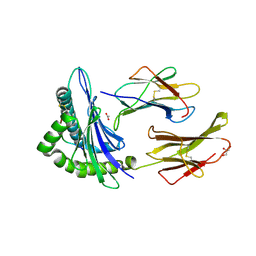 | | Engineered Human HLA_A2 MHC Class I molecule in complex with TCR and SV9 peptide | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Meijers, R, Anjanappa, R, Springer, S, Garcia-Alai, M. | | Deposit date: | 2018-12-04 | | Release date: | 2019-07-24 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-throughput peptide-MHC complex generation and kinetic screenings of TCRs with peptide-receptive HLA-A*02:01 molecules.
Sci Immunol, 4, 2019
|
|
3RTT
 
 | | Human MMP-12 catalytic domain in complex with*(R)-N*-Hydroxy-1-(phenethylsulfonyl)pyrrolidine-2-carboxamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-1-[(2-phenylethyl)sulfonyl]-D-prolinamide, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mori, M, Nativi, C. | | Deposit date: | 2011-05-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Contribution of ligand free energy of solvation to design new potent MMPs inhibitors.
J.Med.Chem., 2012
|
|
3RTS
 
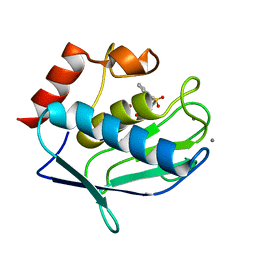 | | Human MMP-12 catalytic domain in complex with*N*-Hydroxy-2-(2-phenylethylsulfonamido)acetamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-N~2~-[(2-phenylethyl)sulfonyl]glycinamide, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mori, M, Nativi, C. | | Deposit date: | 2011-05-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Contribution of ligand free energy of solvation to design new potent MMPs inhibitors.
J.Med.Chem., 2012
|
|
6IB5
 
 | |
6H43
 
 | | Flavin-dependent Tryptophan 6-halogenase Thal | | Descriptor: | GLYCEROL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Moritzer, A, Minges, H, Frese, M, Sewald, N, Niemann, H.H. | | Deposit date: | 2018-07-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based switch of regioselectivity in the flavin-dependent tryptophan 6-halogenase Thal.
J. Biol. Chem., 294, 2019
|
|
6H44
 
 | | Flavin-dependent Tryptophan 6-halogenase Thal in complex with tryptophan | | Descriptor: | GLYCEROL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Moritzer, A, Minges, H, Sewald, N, Niemann, H.H. | | Deposit date: | 2018-07-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-based switch of regioselectivity in the flavin-dependent tryptophan 6-halogenase Thal.
J. Biol. Chem., 294, 2019
|
|
