6TJ1
 
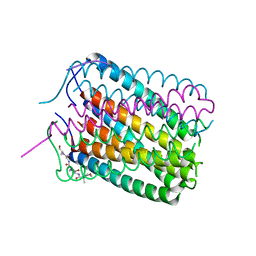 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | Descriptor: | De novo designed WSHC6, purification tag | | Authors: | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | Deposit date: | 2019-11-23 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
1E3H
 
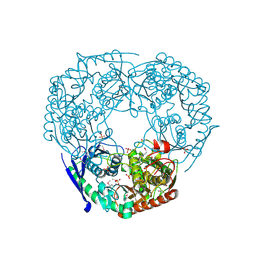 | | SeMet derivative of Streptomyces antibioticus PNPase/GPSI enzyme | | Descriptor: | GUANOSINE PENTAPHOSPHATE SYNTHETASE, SULFATE ION | | Authors: | Symmons, M.F, Jones, G.H, Luisi, B.F. | | Deposit date: | 2000-06-15 | | Release date: | 2000-11-05 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Duplicated Fold is the Structural Basis for Polynucleotide Phosphorylase Catalytic Activity, Processivity, and Regulation
Structure, 8, 2000
|
|
1E3P
 
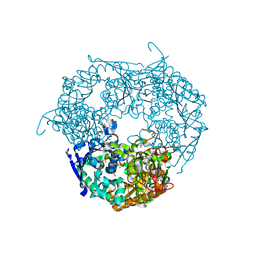 | | tungstate derivative of Streptomyces antibioticus PNPase/GPSI enzyme | | Descriptor: | Polyribonucleotide nucleotidyltransferase, SULFATE ION, TUNGSTATE(VI)ION | | Authors: | Symmons, M.F, Jones, G.H, Luisi, B.F. | | Deposit date: | 2000-06-20 | | Release date: | 2000-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Duplicated Fold is the Structural Basis for Polynucleotide Phosphorylase Catalytic Activity, Processivity, and Regulation
Structure, 8, 2000
|
|
1D98
 
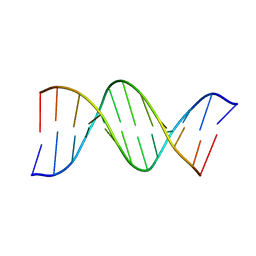 | | THE STRUCTURE OF AN OLIGO(DA).OLIGO(DT) TRACT AND ITS BIOLOGICAL IMPLICATIONS | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Nelson, H.C.M, Finch, J.T, Luisi, B.F, Klug, A. | | Deposit date: | 1992-10-17 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of an oligo(dA).oligo(dT) tract and its biological implications.
Nature, 330, 1987
|
|
8CBK
 
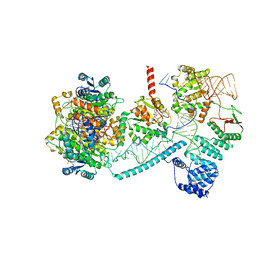 | | Structure of human mitochondrial RNase P in complex with mitochondrial pre-tRNA-His(5,Ser) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, MAGNESIUM ION, Mitochondrial Precursor tRNA-His(5,Ser), ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
8CBL
 
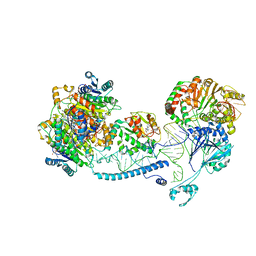 | | Structure of human mitochondrial RNase Z in complex with mitochondrial pre-tRNA-His(0,Ser) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, Mitochondrial Precursor tRNA-His(0,Ser), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, YU, W, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
8CBM
 
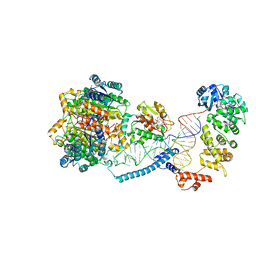 | | Structure of human mitochondrial CCA-adding enzyme in complex with mitochondrial pre-tRNA-Ile | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, CCA tRNA nucleotidyltransferase 1, mitochondrial, ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
8CBO
 
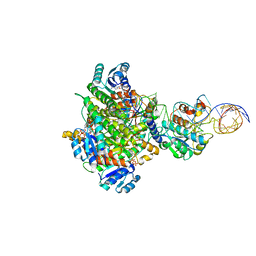 | | Structure of human mitochondrial MRPP1-MRPP2 in complex with mitochondrial pre-tRNA-Ile | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, Mitochondrial Precursor tRNA-Ile(5,4), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
3GME
 
 | |
3GLL
 
 | | Crystal structure of Polynucleotide Phosphorylase (PNPase) core | | Descriptor: | Polyribonucleotide nucleotidyltransferase | | Authors: | Nurmohamed, S, Luisi, B.L. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly.
J.Mol.Biol., 389, 2009
|
|
3GCM
 
 | |
4UOS
 
 | | Thermodynamic hyperstability in parametrically designed helical bundles | | Descriptor: | DESIGNED HELICAL BUNDLE | | Authors: | Oberdorfer, G, Huang, P, Pei, X.Y, Xu, C, Gonen, T, Nannenga, B, DiMaio, D, Rogers, J, Luisi, B.F, Baker, D. | | Deposit date: | 2014-06-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | High Thermodynamic Stability of Parametrically Designed Helical Bundles
Science, 346, 2014
|
|
4UOT
 
 | | Thermodynamic hyperstability in parametrically designed helical bundles | | Descriptor: | DESIGNED HELICAL BUNDLE 5H2L | | Authors: | Oberdorfer, G, Huang, P, Pei, X.Y, Xu, C, Gonen, T, Nannenga, B, DiMaio, D, Rogers, J, Luisi, B.F, Baker, D. | | Deposit date: | 2014-06-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High Thermodynamic Stability of Parametrically Designed Helical Bundles
Science, 346, 2014
|
|
4OXP
 
 | |
5NC5
 
 | |
5NBB
 
 | | Structure of the C-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
5NB9
 
 | | Structure of the N-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
6TPQ
 
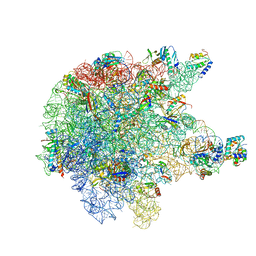 | | RNase M5 bound to 50S ribosome with precursor 5S rRNA | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
6TNN
 
 | | Mini-RNase III (Mini-III) bound to 50S ribosome with precursor 23S rRNA | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | Deposit date: | 2019-12-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
3H1C
 
 | |
287D
 
 | |
8CWM
 
 | |
8CVI
 
 | |
8CXM
 
 | | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | | Descriptor: | Flagellin | | Authors: | Sonani, R.R, Kreutzberger, M.A.B, Sebastian, A.L, Scharf, B, Egelman, E.H. | | Deposit date: | 2022-05-21 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergent evolution in the supercoiling of prokaryotic flagellar filaments.
Cell, 185, 2022
|
|
8CYE
 
 | |
