7VN3
 
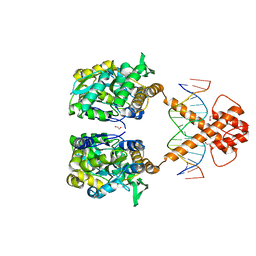 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning CACACGTGTG | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN4
 
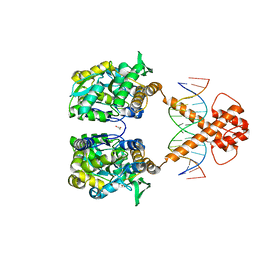 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning TCCACGTGGA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN5
 
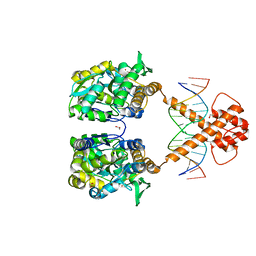 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning TTCACGTGAA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*TP*CP*AP*CP*GP*TP*GP*AP*AP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
6SQL
 
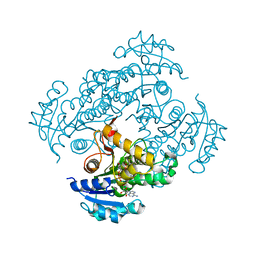 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and N-(3-(aminomethyl)phenyl)-5-chloro-3-methylbenzo[b]thiophene-2-sulfonamide | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ~{N}-[3-(aminomethyl)phenyl]-5-chloranyl-3-methyl-1-benzothiophene-2-sulfonamide | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6SQB
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 3-(3-chlorophenyl)propanoic acid | | Descriptor: | 3-(3-chlorophenyl)propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6SQD
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 2-pyrazol-1-ylbenzoic acid | | Descriptor: | 2-pyrazol-1-ylbenzoic acid, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6SQ9
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 3-hydroxynaphthalene-2-carboxylic acid | | Descriptor: | 3-hydroxynaphthalene-2-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6SQ5
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 3-[3-(trifluoromethyl)phenyl]prop-2-enoic acid | | Descriptor: | (~{E})-3-[3-(trifluoromethyl)phenyl]prop-2-enoic acid, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6BYU
 
 | | X-ray crystal structure of Escherichia coli RNA polymerase (RpoB-H526Y) and ppApp complex | | Descriptor: | (5R)-5-(6-amino-9H-purin-9-yl)-2-({[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}methyl)-4-oxo-4,5-dihydrofuran-3-yl trihydrogen diphosphate, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Murakami, K.S, Molodtsov, V. | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-function comparisons of (p)ppApp vs (p)ppGpp for Escherichia coli RNA polymerase binding sites and for rrnB P1 promoter regulatory responses in vitro.
Biochim. Biophys. Acta, 1861, 2018
|
|
6SQ7
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 2-(4-chloro-3-nitrobenzoyl)benzoic acid | | Descriptor: | 2-(4-chloranyl-3-nitro-phenyl)carbonylbenzoic acid, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
2GEK
 
 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
2WZV
 
 | | Crystal structure of the FMN-dependent nitroreductase NfnB from Mycobacterium smegmatis | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, NFNB PROTEIN, ... | | Authors: | Bellinzoni, M, Manina, G, Riccardi, G, Alzari, P.M. | | Deposit date: | 2009-12-03 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biological and Structural Characterization of the Mycobacterium Smegmatis Nitroreductase Nfnb, and its Role in Benzothiazinone Resistance
Mol.Microbiol., 77, 2010
|
|
2WZW
 
 | | Crystal structure of the FMN-dependent nitroreductase NfnB from Mycobacterium smegmatis in complex with NADPH | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NFNB PROTEIN, ... | | Authors: | Bellinzoni, M, Manina, G, Riccardi, G, Alzari, P.M. | | Deposit date: | 2009-12-03 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biological and Structural Characterization of the Mycobacterium Smegmatis Nitroreductase Nfnb, and its Role in Benzothiazinone Resistance
Mol.Microbiol., 77, 2010
|
|
2DUO
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+-bound form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Vesicular integral-membrane protein VIP36 | | Authors: | Satoh, T, Cowieson, N.P, Kato, R, Wakatsuki, S. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
2DUR
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+/Man2-bound form | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Satoh, T, Cowieson, N.P, Kato, R, Wakatsuki, S. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
2DUQ
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+/Man-bound form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Vesicular integral-membrane protein VIP36, ... | | Authors: | Satoh, T, Cowieson, N.P, Kato, R, Wakatsuki, S. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
2DUP
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, metal-free form | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Satoh, T, Cowieson, N.P, Kato, R, Wakatsuki, S. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
2E6V
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+/Man3GlcNAc-bound form | | Descriptor: | CALCIUM ION, Vesicular integral-membrane protein VIP36, alpha-D-mannopyranose, ... | | Authors: | Satoh, T, Kato, R, Wakatsuki, S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
8IE6
 
 | | Crystal structure of DAPK1 in complex with pinostilbene | | Descriptor: | 3-[(E)-2-(4-hydroxyphenyl)ethenyl]-5-methoxy-phenol, Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
8IE5
 
 | | Crystal structure of DAPK1 in complex with oxyresveratrol | | Descriptor: | Death-associated protein kinase 1, SULFATE ION, trans-oxyresveratrol | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
8IE8
 
 | | Crystal structure of DAPK1 in complex with isorhapontigenin | | Descriptor: | 5-[(~{E})-2-(3-methoxy-4-oxidanyl-phenyl)ethenyl]benzene-1,3-diol, Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
8IE7
 
 | | Crystal structure of DAPK1 in complex with pterostilbene | | Descriptor: | Death-associated protein kinase 1, Pterostilbene, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
8IG1
 
 | | Crystal structure of wild-type transthyretin in complex with rafoxanide | | Descriptor: | SODIUM ION, Transthyretin, ~{N}-[3-chloranyl-4-(4-chloranylphenoxy)phenyl]-3,5-bis(iodanyl)-2-oxidanyl-benzamide | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-20 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Rafoxanide, a salicylanilide anthelmintic, interacts with human plasma protein transthyretin.
Febs J., 290, 2023
|
|
4JK1
 
 | |
4JK2
 
 | |
