2Y80
 
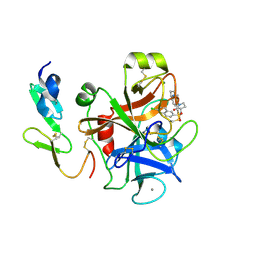 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-{(3S)-1-[(1S)-1-(DIMETHYLAMINO)-2,3-DIHYDRO-1H-INDEN-5-YL]-2-OXO-3-PYRROLIDINYL}-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2WYG
 
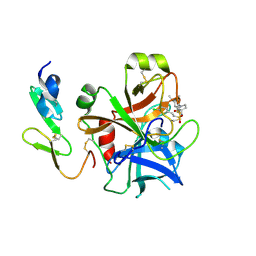 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with monoaryl P4 motifs | | Descriptor: | (E)-2-(5-CHLOROTHIOPHEN-2-YL)-N-[(3S)-1-{4-[(1R)-1-(DIMETHYLAMINO)ETHYL]-2-FLUOROPHENYL}-2-OXOPYRROLIDIN-3-YL]ETHENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN | | Authors: | Kleanthous, S, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chaudry, L, Chan, C, Clarte, M, Convery, M.A, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Roethka, T.J, Senger, S, Shah, G.P, Stelman, G.J, Toomey, J.R, Watson, N.S, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Monoaryl P4 Motifs
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2Y7X
 
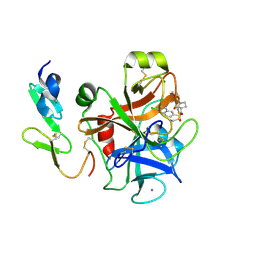 | | The discovery of potent and long-acting oral factor Xa inhibitors with tetrahydroisoquinoline and benzazepine P4 motifs | | Descriptor: | 6-CHLORO-N-[(3S)-1-(5-FLUORO-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL)-2-OXO-PYRROLIDIN-3-YL]NAPHTHALENE-2-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Watson, N.S, Adams, C, Belton, D, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Harling, J.D, Irving, W.R, Irvine, S, Kleanthous, S, McLay, I.M, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Discovery of Potent and Long-Acting Oral Factor Xa Inhibitors with Tetrahydroisoquinoline and Benzazepine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2MBB
 
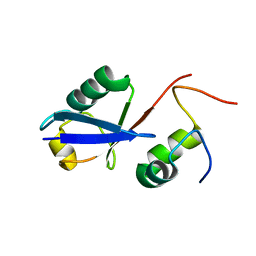 | | Solution Structure of the human Polymerase iota UBM1-Ubiquitin Complex | | Descriptor: | Immunoglobulin G-binding protein G/DNA polymerase iota fusion protein, Polyubiquitin-B | | Authors: | Wang, S, Zhou, P. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sparsely-sampled, high-resolution 4-D omit spectra for detection and assignment of intermolecular NOEs of protein complexes.
J.Biomol.Nmr, 59, 2014
|
|
2WYJ
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with monoaryl P4 motifs | | Descriptor: | (E)-2-(5-CHLOROTHIOPHEN-2-YL)-N-[(3S)-1-{4-[(1S)-1-(DIMETHYLAMINO)ETHYL]-2-FLUOROPHENYL}-2-OXOPYRROLIDIN-3-YL]ETHENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN | | Authors: | Kleanthous, S, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chaudry, L, Chan, C, Clarte, M, Convery, M.A, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Roethka, T.J, Senger, S, Shah, G.P, Stelman, G.J, Toomey, J.R, Watson, N.S, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Monoaryl P4 Motifs
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2MUR
 
 | |
2MUQ
 
 | | Solution Structure of the Human FAAP20 UBZ | | Descriptor: | Fanconi anemia-associated protein of 20 kDa, ZINC ION | | Authors: | Wojtaszek, J.L, Wang, S, Zhou, P. | | Deposit date: | 2014-09-16 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin recognition by FAAP20 expands the complex interface beyond the canonical UBZ domain.
Nucleic Acids Res., 42, 2014
|
|
2Y7Z
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-[(3S)-1-[(1S)-1-DIMETHYLAMINO-2,3-DIHYDRO-1H-INDEN-5-YL]-2-OXO-PYRROLIDIN-3-YL]NAPHTHALENE-2-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y81
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-((3S)-2-OXO-1-{4-[(2R)-2--PYRROLIDINYL] PHENYL}-3-PYRROLIDINYL)-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2MRM
 
 | |
2JT2
 
 | | Solution Structure of the Aquifex aeolicus LpxC- CHIR-090 complex | | Descriptor: | N-{(1S,2R)-2-hydroxy-1-[(hydroxyamino)carbonyl]propyl}-4-{[4-(morpholin-4-ylmethyl)phenyl]ethynyl}benzamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Barb, A.W, Jiang, L, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2007-07-18 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the deacetylase LpxC bound to the antibiotic CHIR-090: Time-dependent inhibition and specificity in ligand binding
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2LSJ
 
 | |
2LSG
 
 | |
1ZGW
 
 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
1XXE
 
 | | RDC refined solution structure of the AaLpxC/TU-514 complex | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Coggins, B.E, McClerren, A.L, Jiang, L, Li, X, Rudolph, J, Hindsgaul, O, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the LpxC-TU-514 Complex and pK(a) Analysis of an Active Site Histidine: Insights into the Mechanism and Inhibitor Design
Biochemistry, 44, 2005
|
|
1U8B
 
 | | Crystal structure of the methylated N-ADA/DNA complex | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*T)-3', 5'-D(*TP*AP*AP*AP*TP*T)-3', 5'-D(P*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*AP*T)-3', ... | | Authors: | He, C, Hus, J.-C, Sun, L.J, Zhou, P, Norman, D.P.G, Dotsch, V, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2004-08-05 | | Release date: | 2005-10-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada.
Mol.Cell, 20, 2005
|
|
1NFA
 
 | | HUMAN TRANSCRIPTION FACTOR NFATC DNA BINDING DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | HUMAN TRANSCRIPTION FACTOR NFATC1 | | Authors: | Wolfe, S.A, Zhou, P, Dotsch, V, Chen, L, You, A, Ho, S.N, Crabtree, G.R, Wagner, G, Verdine, G.L. | | Deposit date: | 1997-01-18 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Unusual Rel-like architecture in the DNA-binding domain of the transcription factor NFATc.
Nature, 385, 1997
|
|
2B5L
 
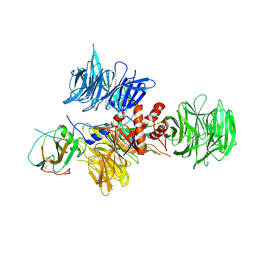 | | Crystal Structure of DDB1 In Complex with Simian Virus 5 V Protein | | Descriptor: | Nonstructural protein V, ZINC ION, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
2B5M
 
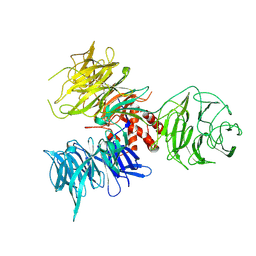 | | Crystal Structure of DDB1 | | Descriptor: | damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
2A7O
 
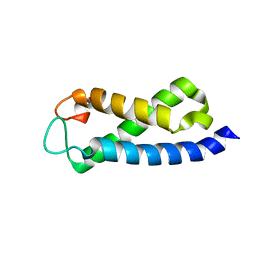 | | Solution Structure of the hSet2/HYPB SRI domain | | Descriptor: | Huntingtin interacting protein B | | Authors: | Li, M, Phatnani, H.P, Guan, Z, Sage, H, Greenleaf, A, Zhou, P. | | Deposit date: | 2005-07-05 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Set2 Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2B5N
 
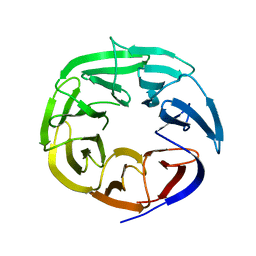 | | Crystal Structure of the DDB1 BPB Domain | | Descriptor: | ISOPROPYL ALCOHOL, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
2BRR
 
 | | Complex of the neisserial PorA P1.4 epitope peptide and two Fab- fragments (antibody MN20B9.34) | | Descriptor: | ACETIC ACID, CLASS 1 OUTER MEMBRANE PROTEIN VARIABLE REGION 2, MN20B9.34 ANTI-P1.4 ANTIBODY, ... | | Authors: | Oomen, C.J, Hoogerhout, P, Kuipers, B, Vidarsson, G, Van Alphen, L, Gros, P. | | Deposit date: | 2005-05-11 | | Release date: | 2005-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Anti-Meningococcal Subtype P1.4 Pora Antibody Provides Basis for Peptide- Vaccine Design.
J.Mol.Biol., 351, 2005
|
|
5XAF
 
 | | Crystal structure of tubulin-stathmin-TTL-Compound Z1 complex | | Descriptor: | (3S,4R)-4-(3-hydroxy-4-methoxyphenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
5XAG
 
 | | Crystal structure of tubulin-stathmin-TTL-Compound Z2 complex | | Descriptor: | (3~{R},4~{R})-3-(hydroxymethyl)-4-(4-methoxy-3-oxidanyl-phenyl)-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
7OX8
 
 | | Target-bound SpCas9 complex with TRAC full RNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
