5MA3
 
 | | GFP-binding DARPin fusion gc_R11 | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, R11 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MFD
 
 | | Designed armadillo repeat protein YIIIM''6AII in complex with pD_(KR)5 | | Descriptor: | CALCIUM ION, Capsid decoration protein,pD_(KR)5, YIIIM''6AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
5MA4
 
 | | GFP-binding DARPin fusion gc_K7 | | Descriptor: | Green fluorescent protein, K7 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MFE
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with (RR)4 peptide | | Descriptor: | (RR)4, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFM
 
 | | Designed armadillo repeat protein peptide fusion YIIIM6AII_GS11_(KR)5 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Importin subunit alpha, ... | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MA5
 
 | | GFP-binding DARPin fusion gc_K11 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MFC
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with (KR)4-GFP | | Descriptor: | (KR)4-Green fluorescent protein,Green fluorescent protein, ACETATE ION, YIIIM5AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
5MFL
 
 | | Designed armadillo repeat protein (KR)5_GS10_YIIIM6AII | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (KR)5_GS10_YIIIM6AII, 1,2-ETHANEDIOL, ... | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFI
 
 | | Designed armadillo repeat protein YIII(Dq.V2)4CqI in complex with peptide (KR)4 | | Descriptor: | (KR)4, YIII(Dq.V2)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFK
 
 | | Designed armadillo repeat protein YIII(Dq.V1)4CPAF in complex with peptide (KR)4 | | Descriptor: | (KR)4, YIII(Dq.V1)4CPAF | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFF
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)5 | | Descriptor: | (RR)5, 1,2-ETHANEDIOL, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFN
 
 | | Designed armadillo repeat protein YIIIM5AII | | Descriptor: | CALCIUM ION, D-MALATE, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MAD
 
 | | GFP-binding DARPin 3G61 | | Descriptor: | 3G61, DI(HYDROXYETHYL)ETHER, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MFB
 
 | | Designed armadillo repeat protein YIII(Dq)4CqI | | Descriptor: | YIII(Dq)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFH
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)5 | | Descriptor: | (RR)5, CALCIUM ION, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MA9
 
 | | GFP-binding DARPin fusion gc_R11 | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, R11 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MFG
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)4 | | Descriptor: | (RR)4, CALCIUM ION, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5XVK
 
 | | Crystal structure of mouse Nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl Nicotinamide (MNA) | | Descriptor: | 3-carbamoyl-1-methylpyridin-1-ium, GLYCEROL, Nicotinamide N-methyltransferase, ... | | Authors: | Swaminathan, S, Birudukota, S, Thakur, M.K, Parveen, R, Kandan, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of monkey and mouse nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl nicotinamide
Biochem. Biophys. Res. Commun., 491, 2017
|
|
1HZ3
 
 | | ALZHEIMER'S DISEASE AMYLOID-BETA PEPTIDE (RESIDUES 10-35) | | Descriptor: | A-BETA AMYLOID | | Authors: | Zhang, S, Iwata, K, Lachenmann, M.J, Peng, J.W, Li, S, Stimson, E.R, Lu, Y, Felix, A.M, Maggio, J.E, Lee, J.P. | | Deposit date: | 2001-01-23 | | Release date: | 2001-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Alzheimer's peptide a beta adopts a collapsed coil structure in water.
J.Struct.Biol., 130, 2000
|
|
6LKB
 
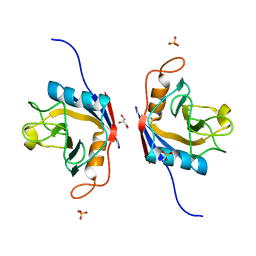 | | Crystal Structure of the peptidylprolyl isomerase domain of Arabidopsis thaliana CYP71. | | Descriptor: | COBALT (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lakhanpal, S, Jobichen, C, Swaminathan, K. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analyses of the PPIase domain of Arabidopsis thaliana CYP71 reveal its catalytic activity toward histone H3.
Febs Lett., 595, 2021
|
|
6SJU
 
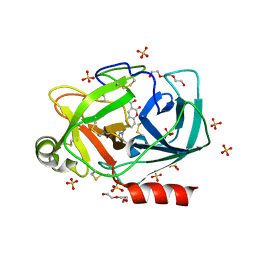 | |
6SHI
 
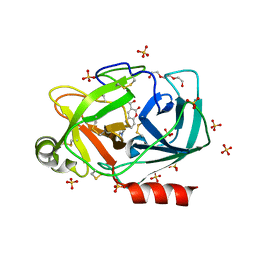 | |
5XYN
 
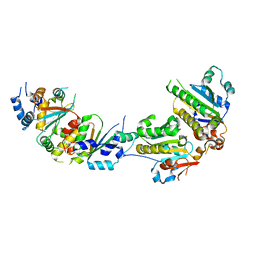 | | The crystal structure of Csm2-Psy3-Shu1-Shu2 complex from budding yeast | | Descriptor: | Chromosome segregation in meiosis protein 2, Platinum sensitivity protein 3, Suppressor of HU sensitivity involved in recombination protein 1, ... | | Authors: | Zhang, S, Zhang, T, Ding, J. | | Deposit date: | 2017-07-09 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the functional role of the Shu complex in homologous recombination.
Nucleic Acids Res., 45, 2017
|
|
6SHH
 
 | |
5H4D
 
 | | Crystal structure of hSIRT3 in complex with a specific agonist Amiodarone hydrochloride | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 7-AMINO-4-METHYL-CHROMEN-2-ONE, ARG-HIS-LYS, ... | | Authors: | Zhang, S, Fu, L, Liu, J, Liu, B. | | Deposit date: | 2016-10-31 | | Release date: | 2017-11-08 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of hSIRT3 in complex with a specific agonist Amiodarone hydrochloride
To Be Published
|
|
