6Z1J
 
 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV LSP co-crystallization with spheroidene | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A.G, Fufina, T.Y, Vasilieva, L.G, Betzel, C, Selikhanov, G.K. | | Deposit date: | 2020-05-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel approaches for the lipid sponge phase crystallization of the Rhodobacter sphaeroides photosynthetic reaction center.
Iucrj, 7, 2020
|
|
2V6W
 
 | | tRNASer acceptor stem: Conformation and hydration of a microhelix in a crystal structure at 1.8 Angstrom resolution | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*AP)-3', 5'-R(*UP*CP*UP*CP*UP*CP*CP)-3' | | Authors: | Foerster, C, Brauer, A.B.E, Brode, S, Fuerste, J.P, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-07-23 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trnaser Acceptor Stem: Conformation and Hydration of a Microhelix in a Crystal Structure at 1.8 A Resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5EP8
 
 | | X-Ray Structure of the Complex Pyrimidine-nucleoside phosphorylase from Bacillus subtilis with Sulfate Ion | | Descriptor: | Pyrimidine-nucleoside phosphorylase, SODIUM ION, SULFATE ION | | Authors: | Lashkov, A.A, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | X-Ray Structure of the Complex Pyrimidine-nucleoside phosphorylase from Bacillus subtilis with Sulfate Ion
To Be Published
|
|
5EPU
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with cytosine at 1.06A. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with cytosine at 1.06A.
To Be Published
|
|
2W89
 
 | | Crystal structure of the E.coli tRNAArg aminoacyl stem issoacceptor RR-1660 at 2.0 Angstroem resolution | | Descriptor: | 5'-R(*CP*GP*GP*AP*UP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*CP*CP*GP)-3', GLYCEROL | | Authors: | Eichert, A, Schreiber, A, Fuerste, J.P, Perbandt, M, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2009-01-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the E. Coli tRNA(Arg) Aminoacyl Stem Isoacceptor Rr-1660 at 2.0 A Resolution.
Biochem.Biophys.Res.Commun., 385, 2009
|
|
2X2Q
 
 | | Crystal structure of an 'all locked' LNA duplex at 1.9 angstrom resolution | | Descriptor: | CACODYLATE ION, COBALT HEXAMMINE(III), LOCKED NUCLEIC ACID DERIVED FROM TRNA SER ACCEPTOR STEM MICROHELIX, ... | | Authors: | Eichert, A, Behling, K, Fuerste, J.P, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2010-01-15 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of an 'All Locked' Nucleic Acid Duplex.
Nucleic Acids Res., 38, 2010
|
|
2VUQ
 
 | | Crystal structure of a human tRNAGly acceptor stem microhelix (derived from the gene sequence DG9990) at 1.18 Angstroem resolution | | Descriptor: | 5'-R(*CP*CP*AP*AP*UP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*UP*GP*GP)-3' | | Authors: | Eichert, A, Perbandt, M, Schreiber, A, Fuerste, J.P, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2008-05-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal Structure of the Human Trnagly Microhelix Isoacceptor G9990 at 1.18 A Resolution
Biochem.Biophys.Res.Commun., 380, 2009
|
|
2VAL
 
 | | Crystal structure of an Escherichia coli tRNAGly microhelix at 2.0 Angstrom resolution | | Descriptor: | 5'-R(*GP*CP*GP*GP*GP*AP*AP)-3', 5'-R(*UP*UP*CP*CP*CP*GP*CP)-3', MAGNESIUM ION | | Authors: | Forster, C, Brauer, A.B.E, Perbandt, M, Lehmann, D, Furste, J.P, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of an Escherichia Coli Trnagly Microhelix at 2.0 Angstrom Resolution
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2V7R
 
 | | Crystal structure of a human tRNAGly microhelix at 1.2 Angstrom resolution | | Descriptor: | HUMAN TRNAGLY MICROHELIX | | Authors: | Foerster, C, Mankowska, M, Fuerste, J.P, Perbandt, M, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-08-01 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of a Human Trnagly Microhelix at 1.2 A Resolution.
Biochem.Biophys.Res.Commun., 368, 2008
|
|
4E0V
 
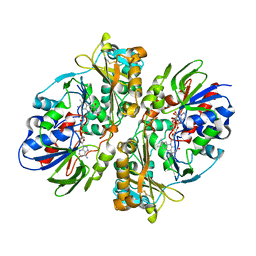 | | Structure of L-amino acid oxidase from the B. jararacussu venom | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-amino-acid oxidase | | Authors: | Ullah, A, Souza, T.A.C.B, Betzel, C, Murakami, M.T, Arni, R.K. | | Deposit date: | 2012-03-05 | | Release date: | 2012-04-25 | | Last modified: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into selectivity and cofactor binding in snake venom L-amino acid oxidases.
Biochem.Biophys.Res.Commun., 421, 2012
|
|
4EB2
 
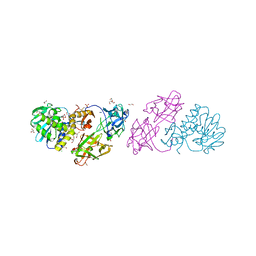 | | Crystal structure Mistletoe Lectin I from Viscum album in complex with n-acetyl-d-glucosamine at 1.94 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Laskov, A.A, Prokofev, I.I, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure Mistletoe Lectin I from Viscum album in complex with N-acetyl-D-glucosamine at 1.94 A resolution.
To be Published
|
|
4E3Y
 
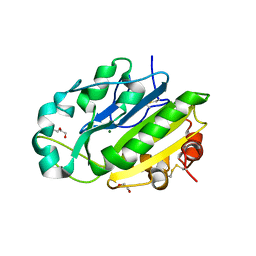 | | X-ray structure of the Serratia marcescens endonuclease at 0.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Lashkov, A.A, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2012-03-11 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-ray structure of the Serratia marcescens endonuclease at 0.95 A resolution
To be Published
|
|
4DCF
 
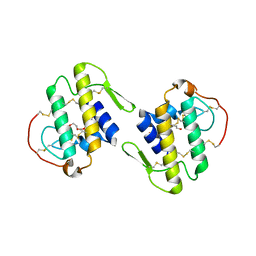 | | Structure of MTX-II from Bothrops brazili | | Descriptor: | MTX-II chain A, TETRAETHYLENE GLYCOL | | Authors: | Ullah, A, Souza, T.A.C.B, Betzel, C, Murakami, M.T, Arni, R.K. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic portrayal of different conformational states of a Lys49 phospholipase A2 homologue: insights into structural determinants for myotoxicity and dimeric configuration.
Int.J.Biol.Macromol., 51, 2012
|
|
6ELY
 
 | | Crystal Structure of Mistletoe Lectin I (ML-I) from Viscum album in Complex with 4-N-Furfurylcytosine at 2.84 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-N-Furfurylcytosine, ... | | Authors: | Ahmad, M.S, Rasheed, S, Falke, S, Khaliq, B, Perbandt, M, Choudhary, M.I, Markiewicz, W.T, Barciszewski, J, Betzel, C. | | Deposit date: | 2017-09-30 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal Structure of Mistletoe Lectin I (ML-I) from Viscum album in Complex with 4-N-Furfurylcytosine at 2.85 angstrom Resolution.
Med Chem, 14, 2018
|
|
6EYP
 
 | | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Prokofev, I.I, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Lashkov, A.A. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A
To Be Published
|
|
2YVV
 
 | | Crystal structure of hyluranidase complexed with lactose at 2.6 A resolution reveals three specific sugar recognition sites | | Descriptor: | Hyaluronidase, phage associated, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mishra, P, Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Bhakuni, V, Singh, T.P. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of hyluranidase complexed with lactose at 2.6 A resolution reveals three specific sugar recognition sites
To be Published
|
|
2YW0
 
 | | Crystal structure of hyluranidase trimer at 2.6 A resolution | | Descriptor: | Hyaluronidase, phage associated | | Authors: | Prem Kumar, R, Mishra, P, Singh, N, Perbandt, M, Kaur, P, Sharma, S, Betzel, C, Bhakuni, V, Singh, T.P. | | Deposit date: | 2007-04-18 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Polysaccharide binding sites in hyaluronate lyase--crystal structures of native phage-encoded hyaluronate lyase and its complexes with ascorbic acid and lactose
Febs J., 276, 2009
|
|
2YX2
 
 | | Crystal structure of cloned trimeric hyluranidase from streptococcus pyogenes at 2.8 A resolution | | Descriptor: | Hyaluronidase, phage associated | | Authors: | Mishra, P, Prem Kumar, R, Bhakuni, V, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cloned trimeric hyluranidase from streptococcus pyogenes at 2.8 A resolution
To be Published
|
|
361D
 
 | | CRYSTAL STRUCTURE OF DOMAIN E OF THERMUS FLAVUS 5S RRNA: A HELICAL RNA-STRUCTURE INCLUDING A TETRALOOP | | Descriptor: | RNA (5'-R(*CP*UP*GP*GP*GP*CP*GP*GP*GP*CP*GP*AP*CP*CP*GP*CP*C P*UP*GP*G)-3') | | Authors: | Perbandt, M, Nolte, A, Lorenz, S, Erdmann, V.A, Betzel, C. | | Deposit date: | 1997-11-10 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of domain E of Thermus flavus 5S rRNA: a helical RNA structure including a hairpin loop.
FEBS Lett., 429, 1998
|
|
1AOK
 
 | | VIPOXIN COMPLEX | | Descriptor: | ACETATE ION, VIPOXIN COMPLEX | | Authors: | Perbandt, M, Wilson, J.C, Eschenburg, S, Betzel, C. | | Deposit date: | 1997-07-07 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of vipoxin at 2.0 A: an example of regulation of a toxic function generated by molecular evolution.
FEBS Lett., 412, 1997
|
|
4QXD
 
 | | Crystal structure of Inositol Polyphosphate 1-Phosphatase from Entamoeba histolytica | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, putative, MAGNESIUM ION, ... | | Authors: | Tarique, K.F, Abdul Rehman, S.A, Betzel, C, Gourinath, S. | | Deposit date: | 2014-07-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-based identification of inositol polyphosphate 1-phosphatase from Entamoeba histolytica
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1JLT
 
 | | Vipoxin Complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHOLIPASE A2, ... | | Authors: | Banumathi, S, Rajashankar, K.R, Notzel, C, Aleksiev, B, Singh, T.P, Genov, N, Betzel, C. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the neurotoxic complex vipoxin at 1.4 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4YYY
 
 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with uridine | | Descriptor: | CITRIC ACID, TRIETHYLENE GLYCOL, Thymidine phosphorylase, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-03-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5C80
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with uridine at 2.24 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-06-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases
Crystallography Reports, 61, 2016
|
|
4ZXG
 
 | | Ligandin binding site of PfGST | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Glutathione S-transferase, ... | | Authors: | Perbandt, M, Eberle, R, Betzel, C. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution structures of Plasmodium falciparum GST complexes provide novel insights into the dimer-tetramer transition and a novel ligand-binding site.
J.Struct.Biol., 191, 2015
|
|
