6P70
 
 | | X-ray crystal structure of bacterial RNA polymerase and pyrBI promoter complex | | Descriptor: | DNA (5'-D(*TP*AP*TP*AP*AP*TP*CP*GP*AP*TP*CP*TP*TP*TP*GP*CP*CP*GP*GP*G)-3'), DNA (5'-D(P*TP*CP*CP*CP*GP*GP*CP*AP*AP*AP*TP*TP*GP*TP*CP*CP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shin, Y, Murakami, K.S. | | Deposit date: | 2019-06-04 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Structural basis of reiterative transcription from the pyrG and pyrBI promoters by bacterial RNA polymerase.
Nucleic Acids Res., 48, 2020
|
|
6P71
 
 | |
7A1O
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 4-ethyl-2-oxoglutarate, AND CONSENSUS ANKYRIN REPEAT DOMAIN (20-MER) | | Descriptor: | 4-ethyl-2-oxoglutarate, CONSENSUS ANKYRIN REPEAT DOMAIN, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1Q
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 3-(carboxycarbonyl)cyclopentane-1-carboxylic acid, AND CONSENSUS ANKYRIN REPEAT DOMAIN (20-MER) | | Descriptor: | 3-(carboxycarbonyl)cyclopentane-1-carboxylic acid, CONSENSUS ANKYRIN REPEAT DOMAIN, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1P
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 4-propyl-2-oxoglutarate, AND CONSENSUS ANKYRIN REPEAT DOMAIN (20-MER) | | Descriptor: | (4~{S})-2-oxidanylidene-4-propyl-pentanedioic acid, CONSENSUS ANKYRIN REPEAT DOMAIN, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1M
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II) AND 3-propyl-2-oxoglutarate | | Descriptor: | 3-propyl-2-oxoglutarate, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1N
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 3-methyl-2-oxoglutarate, AND CONSENSUS ANKYRIN REPEAT DOMAIN (20-MER) | | Descriptor: | (3~{S})-3-methyl-2-oxidanylidene-pentanedioic acid, CONSENSUS ANKYRIN REPEAT DOMAIN, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1L
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II) AND 3-methyl-2-oxoglutarate | | Descriptor: | (3~{S})-3-methyl-2-oxidanylidene-pentanedioic acid, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1J
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II) AND 3-(3-phenylpropyl)-2-oxoglutarate | | Descriptor: | 3-(3-phenylpropyl)-2-oxoglutarate, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1S
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 3-methyl-2-oxoglutarate, AND TANKYRASE-2 (TNKS2) FRAGMENT PEPTIDE (21-MER) | | Descriptor: | (3~{S})-3-methyl-2-oxidanylidene-pentanedioic acid, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1K
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II) AND 3-((9,9-dimethyl-9H-fluoren-2-yl)methyl)-2-oxoglutarate | | Descriptor: | 3-((9,9-dimethyl-9H-fluoren-2-yl)methyl)-2-oxoglutarate, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7BOM
 
 | | Crystal structure of recombinant horse spleen apo-R52C/E56C/R59C/E63C-Fr immobilized with gold ions. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Maity, B, Ito, N, Abe, S, Lu, D, Ueno, T. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design of Multinuclear Gold Binding Site at the Two-fold Symmetric Interface of the Ferritin Cage
Chem Lett., 49, 2021
|
|
7BON
 
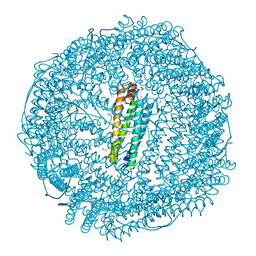 | | Crystal structure of recombinant horse spleen apo-R52C/E56C/R59C/E63C-Fr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Maity, B, Ito, N, Abe, S, Lu, D, Ueno, T. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Design of Multinuclear Gold Binding Site at the Two-fold Symmetric Interface of the Ferritin Cage
Chem Lett., 49, 2021
|
|
5KC1
 
 | | Structure of the C-terminal dimerization domain of Atg38 | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Autophagy-related protein 38, ... | | Authors: | Ohashi, Y, Soler, N, Garcia-Ortegon, M, Zhang, L, Perisic, O, Masson, G.R, Johnson, C.M, Williams, R.J. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
7YBC
 
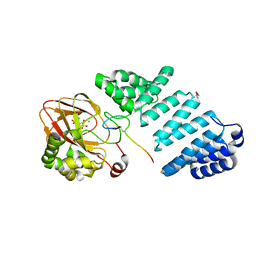 | |
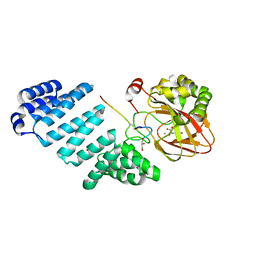
7YB8
 
 | |
7YBA
 
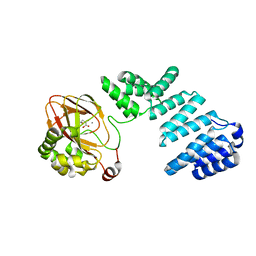 | |
7YB9
 
 | |
7YBB
 
 | |
4ZID
 
 | | Dimeric Hydrogenobacter thermophilus cytochrome c552 obtained from Escherichia coli | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Hayashi, Y, Yamanaka, M, Nagao, S, Komori, H, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-04-28 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Domain swapping oligomerization of thermostable c-type cytochrome in E. coli cells
Sci Rep, 6, 2016
|
|
7R8I
 
 | |
5HEE
 
 | | Crystal structure of the TK2203 protein | | Descriptor: | GLYCEROL, Putative uncharacterized protein, TK2203 protein, ... | | Authors: | Nishitani, Y, Miki, K. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of the TK2203 protein from Thermococcus kodakarensis, a putative extradiol dioxygenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3O26
 
 | |
1JDC
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 1) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1JDD
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 2) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
