8V5H
 
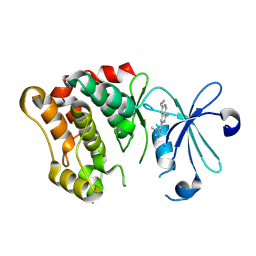 | |
8V5I
 
 | | Crystal structure of MAP4K4 in complex with an inhibitor | | Descriptor: | (3M)-N~6~-(1,4-dimethyl-1H-pyrazol-3-yl)-3-(1-methyl-1H-imidazol-5-yl)-2,7-naphthyridine-1,6-diamine, Mitogen-activated protein kinase kinase kinase kinase 4 | | Authors: | Greasley, S.E, Diehl, W. | | Deposit date: | 2023-11-30 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Microtubule-Associated Serine/Threonine Kinase-like (MASTL).
J.Med.Chem., 67, 2024
|
|
9CZT
 
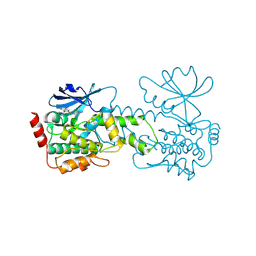 | | HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with compound 6 | | Descriptor: | 4-(aminomethyl)-2-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}-2,3-dihydro-1H-isoindol-1-one, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Johnson, E, Mc Tigue, M. | | Deposit date: | 2024-08-05 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of PF-07265028, A Selective Small Molecule Inhibitor of Hematopoietic Progenitor Kinase 1 (HPK1) for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
9CZX
 
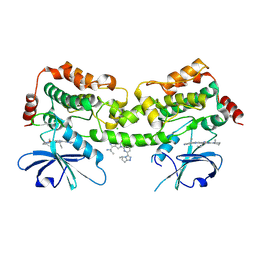 | | HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with compound 21 | | Descriptor: | 4-[(1R)-1-aminopropyl]-2-{6-[(4S,5S)-5-methyl-6,7-dihydro-5H-pyrrolo[2,1-c][1,2,4]triazol-3-yl]pyridin-2-yl}-6-[(2R)-2-methylpyrrolidin-1-yl]-2,3-dihydro-1H-pyrrolo[3,4-c]pyridin-1-one, CHLORIDE ION, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Johnson, E, Mc Tigue, M. | | Deposit date: | 2024-08-05 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.464 Å) | | Cite: | Discovery of PF-07265028, A Selective Small Molecule Inhibitor of Hematopoietic Progenitor Kinase 1 (HPK1) for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
9CZU
 
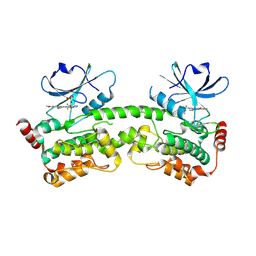 | | HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with compound 9 | | Descriptor: | 6-methoxy-4-[(methylamino)methyl]-2-(6-{4-[(2S)-4,4,4-trifluorobutan-2-yl]-4H-1,2,4-triazol-3-yl}pyridin-2-yl)-2,3-dihydro-1H-isoindol-1-one, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | Johnson, E, Mc Tigue, M. | | Deposit date: | 2024-08-05 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of PF-07265028, A Selective Small Molecule Inhibitor of Hematopoietic Progenitor Kinase 1 (HPK1) for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
9CZW
 
 | | HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with compound 13 | | Descriptor: | 6-(dimethylamino)-4-[(methylamino)methyl]-2-[6-(4-propyl-4H-1,2,4-triazol-3-yl)pyridin-2-yl]-2,3-dihydro-1H-pyrrolo[3,4-c]pyridin-1-one, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Johnson, E, Mc Tigue, M. | | Deposit date: | 2024-08-05 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of PF-07265028, A Selective Small Molecule Inhibitor of Hematopoietic Progenitor Kinase 1 (HPK1) for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
9D00
 
 | | HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with compound 53 | | Descriptor: | 4-[(1R)-1-aminopropyl]-6-methoxy-2-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}-2,3-dihydro-1H-isoindol-1-one, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Johnson, E, McTigue, M, Cronin, C.N. | | Deposit date: | 2024-08-05 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Discovery of PF-07265028, A Selective Small Molecule Inhibitor of Hematopoietic Progenitor Kinase 1 (HPK1) for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
4MUW
 
 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | Descriptor: | 2-{4-[(6,7-difluoro-1H-benzimidazol-2-yl)amino]phenoxy}-N-methyl-3,4'-bipyridin-2'-amine, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S, Jordan, S. | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
4MVH
 
 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | Descriptor: | 1H-benzimidazol-2-yl(4-{[3-(morpholin-4-yl)pyrazin-2-yl]oxy}phenyl)methanone, SULFATE ION, ZINC ION, ... | | Authors: | Chmait, S, Jordan, S. | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
7L0N
 
 | | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Snell, G, Czudnochowski, N, Dillen, J, Nix, J.C, Croll, T.I, Corti, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity.
Cell, 184, 2021
|
|
4P0N
 
 | | Crystal structure of PDE10a with a novel Imidazo[4,5-b]pyridine inhibitor | | Descriptor: | GLYCEROL, N-[cis-3-(2-methoxy-3H-imidazo[4,5-b]pyridin-3-yl)cyclobutyl]-1,3-benzothiazol-2-amine, N-[trans-3-(2-methoxy-3H-imidazo[4,5-b]pyridin-3-yl)cyclobutyl]-1,3-benzothiazol-2-amine, ... | | Authors: | Chmait, S. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Novel Imidazo[4,5-b]pyridines as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
Acs Med.Chem.Lett., 5, 2014
|
|
6Z46
 
 | |
4P1R
 
 | |
7JQD
 
 | | Crystal Structure of PAC1r in complex with peptide antagonist | | Descriptor: | Peptide-43, Pituitary adenylate cyclase-activating polypeptide type I receptor | | Authors: | Piper, D.E, Hu, E, Fang-Tsao, H. | | Deposit date: | 2020-08-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Selective Pituitary Adenylate Cyclase 1 Receptor (PAC1R) Antagonist Peptides Potent in a Maxadilan/PACAP38-Induced Increase in Blood Flow Pharmacodynamic Model.
J.Med.Chem., 64, 2021
|
|
4PHW
 
 | |
4TPP
 
 | | 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors | | Descriptor: | 1-[4-(3-{[1-(quinolin-2-yl)azetidin-3-yl]oxy}quinoxalin-2-yl)piperidin-1-yl]ethanone, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S. | | Deposit date: | 2014-06-09 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and preliminary biological evaluation of potent and selective 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors with improved solubility.
Bioorg.Med.Chem., 22, 2014
|
|
4TPM
 
 | |
