2YB4
 
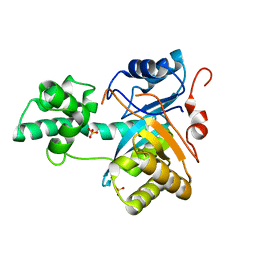 | | Structure of an amidohydrolase from Chromobacterium violaceum (EFI target EFI-500202) with bound SO4, no metal | | Descriptor: | AMIDOHYDROLASE, SULFATE ION | | Authors: | Vetting, M.W, Hillerich, B, Foti, R, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prospecting for Unannotated Enzymes: Discovery of a 3',5'-Nucleotide Bisphosphate Phosphatase within the Amidohydrolase Superfamily.
Biochemistry, 53, 2014
|
|
1MUE
 
 | | Thrombin-Hirugen-L405,426 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(1-OXIDO-2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUOROPHENYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Nantermet, P.G, Selnick, H.G, Isaacs, R.C, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Stranieri, M.T, Cook, J.J, McMasters, D.R, Pellicore, J.M, Pal, S, Wallace, A.A, Clayton, F.C, Bohn, D, Welsh, D.C, Lynch, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pharmacokinetic optimization of 3-amino-6-chloropyrazinone acetamide thrombin inhibitors. Implementation of P3 pyridine N-oxides to deliver an orally bioavailable series containing P1 N-benzylamides.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
2W11
 
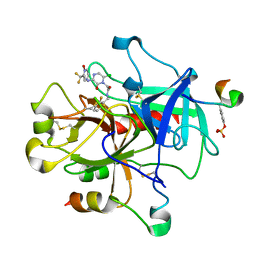
 | | Structure of the L-2-haloacid dehalogenase from Sulfolobus tokodaii | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 2-HALOALKANOIC ACID DEHALOGENASE | | Authors: | Rye, C.A, Isupov, M.N, Lebedev, A.A, Littlechild, J.A. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Studies of a L-Haloacid Dehalogenase from the Thermophilic Archaeon Sulfolobus Tokodaii.
Extremophiles, 13, 2009
|
|
2WKW
 
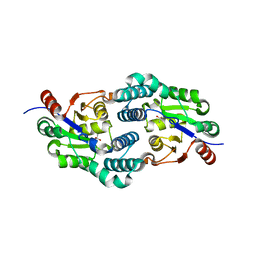
 | | Alcaligenes esterase complexed with product analogue | | Descriptor: | CARBOXYLESTERASE, GLYCEROL, SULFATE ION, ... | | Authors: | Bourne, P.C, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2009-06-18 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Atomic-Resolution Structure of a Novel Bacterial Esterase.
Structure, 8, 2000
|
|
4A2Z
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 4-METHOXY-2,3,6-TRIMETHYL-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
2XME
 
 | | The X-ray structure of CTP:inositol-1-phosphate cytidylyltransferase from Archaeoglobus fulgidus | | Descriptor: | CTP-INOSITOL-1-PHOSPHATE CYTIDYLYLTRANSFERASE, GLYCEROL | | Authors: | Brito, J.A, Borges, N, Vonrhein, C, Santos, H, Archer, M. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Archaeoglobus Fulgidus Ctp:Inositol-1-Phosphate Cytidylyltransferase, a Key Enzyme for Di-Myo-Inositol-Phosphate Synthesis in (Hyper)Thermophiles.
J.Bacteriol., 193, 2011
|
|
2XMH
 
 | | The X-ray structure of CTP:inositol-1-phosphate cytidylyltransferase from Archaeoglobus fulgidus | | Descriptor: | CITRATE ANION, CTP-INOSITOL-1-PHOSPHATE CYTIDYLYLTRANSFERASE | | Authors: | Brito, J.A, Borges, N, Vonrhein, C, Santos, H, Archer, M. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Archaeoglobus fulgidus CTP:inositol-1-phosphate cytidylyltransferase, a key enzyme for di-myo-inositol-phosphate synthesis in (hyper)thermophiles.
J. Bacteriol., 193, 2011
|
|
1MU6
 
 | | Crystal Structure of Thrombin in Complex with L-378,622 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-6-PYRIDINYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Sanderson, P.E, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Williams, P.D, Coburn, C.A, Dorsey, B.D, Barrow, J.C, Stranieri, M.T, Holahan, M.A, Sitko, G.R, Cook, J.J, McMasters, D.R, McDonough, C.M, Sanders, W.M, Wallace, A.A, Clayton, F.C, Bohn, D, Leonard, Y.M, Detwiler Jr, T.J, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P.J. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines.
J.Med.Chem., 46, 2003
|
|
5LTO
 
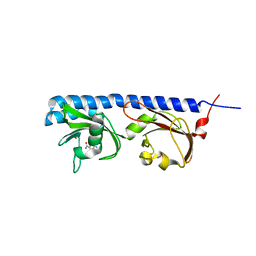 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Gln | | Descriptor: | GLUTAMINE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.459 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LO9
 
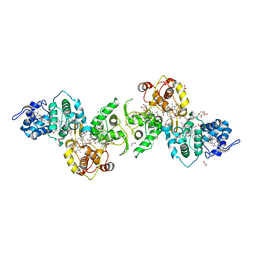 | | Thiosulfate dehydrogenase (TsdBA) from Marichromatium purpuratum - "as isolated" form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytochrome C, ... | | Authors: | Brito, J.A, Kurth, J.M, Reuter, J, Flegler, A, Koch, T, Franke, T, Klein, E, Rowe, S, Butt, J.N, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2016-08-08 | | Release date: | 2016-10-12 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Electron Accepting Units of the Diheme Cytochrome c TsdA, a Bifunctional Thiosulfate Dehydrogenase/Tetrathionate Reductase.
J.Biol.Chem., 291, 2016
|
|
5LT9
 
 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Arg | | Descriptor: | ARGININE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
7S9Z
 
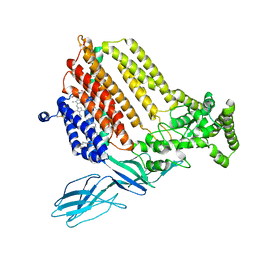 | | Helicobacter Hepaticus CcsBA Closed Conformation | | Descriptor: | Cytochrome c biogenesis protein, HEME B/C, PHOSPHATIDYLETHANOLAMINE | | Authors: | Mendez, D.L, Lowder, E.P, Tillman, D.E, Sutherland, M.C, Collier, A.L, Rau, M.J, Fitzpatrick, J.A, Kranz, R.G. | | Deposit date: | 2021-09-21 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Cryo-EM of CcsBA reveals the basis for cytochrome c biogenesis and heme transport.
Nat.Chem.Biol., 18, 2022
|
|
7S9Y
 
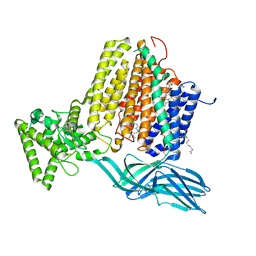 | | Helicobacter Hepaticus CcsBA Open Conformation | | Descriptor: | Cytochrome c biogenesis protein, HEME B/C, PHOSPHATIDYLETHANOLAMINE | | Authors: | Mendez, D.L, Lowder, E.P, Tillman, D.E, Sutherland, M.C, Collier, A.L, Rau, M.J, Fitzpatrick, J.A, Kranz, R.G. | | Deposit date: | 2021-09-21 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM of CcsBA reveals the basis for cytochrome c biogenesis and heme transport.
Nat.Chem.Biol., 18, 2022
|
|
2WKN
 
 | | gamma lactamase from Delftia acidovorans | | Descriptor: | FORMAMIDASE, ZINC ION | | Authors: | Isupov, M.N, Line, K, Gonsalvez, I.S, Gange-Harris, P, Lanzotti, M, Saneei, V, Littlechild, J.A. | | Deposit date: | 2009-06-16 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Structure of Gamma Lactamase from Delftia Acidovorans
To be Published
|
|
1MU8
 
 | | thrombin-hirugen_l-378,650 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-3-METHYL-6-PYRIDINYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Sanderson, P.E, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Williams, P.D, Coburn, C.A, Dorsey, B.D, Barrow, J.C, Stranieri, M.T, Holahan, M.A, Sitko, G.R, Cook, J.J, McMasters, D.R, McDonough, C.M, Sanders, W.M, Wallace, A.A, Clayton, F.C, Bohn, D, Leonard, Y.M, Detwiler Jr, T.J, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P.J. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines.
J.Med.Chem., 46, 2003
|
|
2WMC
 
 | | Crystal structure of eukaryotic initiation factor 4E from Pisum sativum | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | | Authors: | Ashby, J.A, Stevenson, C.E.M, Maule, A.J, Lawson, D.M. | | Deposit date: | 2009-06-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Mutational Analysis of Eif4E in Relation to Sbm1 Resistance to Pea Seed-Borne Mosaic Virus in Pea.
Plos One, 6, 2011
|
|
2YFA
 
 | | X-ray structure of McpS ligand binding domain in complex with malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, ACETATE ION, METHYL-ACCEPTING CHEMOTAXIS TRANSDUCER, ... | | Authors: | Pineda-Molina, E, Gavira, J.A, Krell, T. | | Deposit date: | 2011-04-05 | | Release date: | 2012-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for Chemoreceptors with Bimodular Ligand-Binding Regions Harboring Two Signal-Binding Sites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YB1
 
 | | Structure of an amidohydrolase from Chromobacterium violaceum (EFI target EFI-500202) with bound Mn, AMP and phosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, AMIDOHYDROLASE, MANGANESE (II) ION, ... | | Authors: | Vetting, M.W, Hillerich, B, Foti, R, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Prospecting for Unannotated Enzymes: Discovery of a 3',5'-Nucleotide Bisphosphate Phosphatase within the Amidohydrolase Superfamily.
Biochemistry, 53, 2014
|
|
2YFB
 
 | | X-ray structure of McpS ligand binding domain in complex with succinate | | Descriptor: | ACETATE ION, METHYL-ACCEPTING CHEMOTAXIS TRANSDUCER, SUCCINIC ACID, ... | | Authors: | Gavira, J.A, Pineda-Molina, E, Krell, T. | | Deposit date: | 2011-04-05 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence for Chemoreceptors with Bimodular Ligand-Binding Regions Harboring Two Signal-Binding Sites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3IVK
 
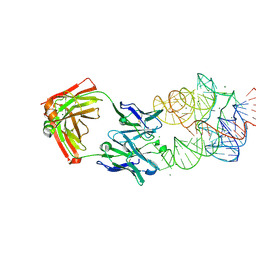 | | Crystal Structure of the Catalytic Core of an RNA Polymerase Ribozyme Complexed with an Antigen Binding Antibody Fragment | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab heavy chain, ... | | Authors: | Koldobskaya, Y, Duguid, E.M, Shechner, D.M, Koide, S, Kossiakoff, A.A, Bartel, D.P, Piccirilli, J.A. | | Deposit date: | 2009-09-01 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
3IZG
 
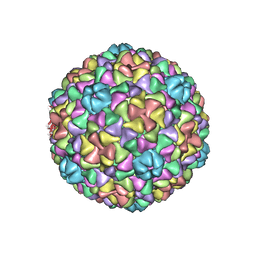 | | Bacteriophage T7 prohead shell EM-derived atomic model | | Descriptor: | Major capsid protein 10A | | Authors: | Ionel, A, Velazquez-Muriel, J.A, Agirrezabala, X, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2010-10-27 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
7JJP
 
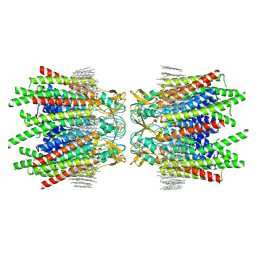 | | Sheep Connexin-50 at 1.9 angstroms resolution by CryoEM | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
5UI9
 
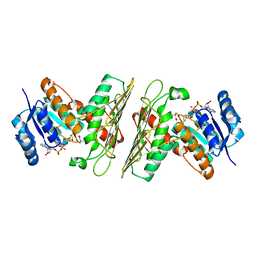 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, 2 -hydroxy-2-hydroxymethyl propanoic acid and Magnesium | | Descriptor: | (2S)-2,3-dihydroxy-2-methylpropanoic acid, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, 2 -hydroxy-2-hydroxymethyl propanoic acid and Magnesium
To be published
|
|
6GCV
 
 | | Ligand binding domain (LBD) of the p. aeruginosa nitrate receptor McpN | | Descriptor: | ACETATE ION, Chemotaxis transducer, NITRATE ION, ... | | Authors: | Gavira, J.A, Krell, T, Martin-Mora, D, Ortega, A. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Molecular Mechanism of Nitrate Chemotaxis via Direct Ligand Binding to the PilJ Domain of McpN.
MBio, 10, 2019
|
|
5UK9
 
 | | Wild-type K-Ras(GCP) pH 6.5 | | Descriptor: | GLYCEROL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Parker, J.A, Mattos, C. | | Deposit date: | 2017-01-20 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | K-Ras Populates Conformational States Differently from Its Isoform H-Ras and Oncogenic Mutant K-RasG12D.
Structure, 26, 2018
|
|
