4UWG
 
 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-2-(morpholin-4-yl)-9-[2-(propan-2-yloxy)ethyl]-8-(trifluoromethyl)-6,7,8,9-tetrahydro-4H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3, SULFATE ION | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxobutyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
4V3B
 
 | | The structure of alpha2,3-sialyltransferase variant 1 from Pasteurella dagmatis in complex with the donor product CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
5K5Y
 
 | | Crystal structure of truncated FlgD (monoclinic form) from the human pathogen Helicobacter pylori (strain 26695) | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Kekez, I, Cendron, L, Stojanovic, M, Zanotti, G, Matkovic-Calogovic, D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Stability of FlgD from the Pathogenic 26695 Strain of Helicobacter pylori
Croatica Chemica Acta, 2016
|
|
4V4X
 
 | | Crystal structure of the 70S Thermus thermophilus ribosome showing how the 16S 3'-end mimicks mRNA E and P codons. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
4V8T
 
 | | Cryo-EM Structure of the 60S Ribosomal Subunit in Complex with Arx1 and Rei1 | | Descriptor: | 25S RIBOSOMAL RNA, 5.8S RIBOSOMAL RNA, 5S RIBOSOMAL RNA, ... | | Authors: | Greber, B.J, Boehringer, D, Montellese, C, Ban, N. | | Deposit date: | 2012-08-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Cryo-Em Structures of Arx1 and Maturation Factors Rei1 and Jjj1 Bound to the 60S Ribosomal Subunit
Nat.Struct.Mol.Biol., 19, 2012
|
|
4V8N
 
 | | The crystal structure of agmatidine tRNA-Ile2 bound to the 70S ribosome in the A and P site. | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Mandal, D, Neubauer, C, Koehrer, C, RajBhandary, U.L, Ramakrishnan, V. | | Deposit date: | 2013-02-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structural Basis for Specific Decoding of Aua by Isoleucine tRNA on the Ribosome
Nat.Struct.Mol.Biol., 20, 2013
|
|
9EWS
 
 | | Optimisation of Potent, Efficacious, Selective and Blood-Brain Barrier Penetrating Inhibitors Targeting EGFR Exon20 Insertion Mutations | | Descriptor: | 1-[3-pyridin-4-yl-2-[3-(2-pyrimidin-2-ylethynyl)phenyl]-4,6-dihydropyrrolo[3,4-d]imidazol-5-yl]propan-1-one, Epidermal growth factor receptor, IODIDE ION | | Authors: | Hargreaves, D. | | Deposit date: | 2024-04-04 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Optimization of Potent, Efficacious, Selective and Blood-Brain Barrier Penetrating Inhibitors Targeting EGFR Exon20 Insertion Mutations.
J.Med.Chem., 2024
|
|
1DBR
 
 | | HYPOXANTHINE GUANINE XANTHINE | | Descriptor: | HYPOXANTHINE GUANINE XANTHINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION | | Authors: | Schumacher, M.A, Carter, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1996-02-13 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Toxoplasma gondii HGXPRTase reveal the catalytic role of a long flexible loop.
Nat.Struct.Biol., 3, 1996
|
|
4W5A
 
 | | Complex structure of ATRX ADD bound to H3K9me3S10ph peptide | | Descriptor: | Peptide from Histone H3.3, Transcriptional regulator ATRX, ZINC ION | | Authors: | Zhao, D, Xiang, B, Li, H. | | Deposit date: | 2014-08-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5JQX
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with phosphoglyceric acid (PGA) - GpgS*PGA | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Glucosyl-3-phosphoglycerate synthase | | Authors: | Albesa-Jove, D, Sancho-Vaello, E, Rodrigo-Unzueta, A, Comino, N, Carreras-Gonzalez, A, Arrasate, P, Urresti, S, Guerin, M.E. | | Deposit date: | 2016-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Snapshots and Loop Dynamics along the Catalytic Cycle of Glycosyltransferase GpgS.
Structure, 25, 2017
|
|
5JVD
 
 | | Tubulin-TUB092 complex | | Descriptor: | (2E)-3-(3-hydroxy-4-methoxyphenyl)-1-(7-methoxy-2H-1,3-benzodioxol-5-yl)-2-methylprop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Canela, M.-D, Noppen, S, Bueno, O, Prota, A.E, Bargsten, K, Saez-Calvo, G, Jimeno, M.-L, Benkheil, M, Ribatti, D, Velazquez, S, Camarasa, M.-J, Diaz, J.F, Steinmetz, M.O, Priego, E.-M, Perez-Perez, M.-J, Liekens, S. | | Deposit date: | 2016-05-11 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Antivascular and antitumor properties of the tubulin-binding chalcone TUB091.
Oncotarget, 8, 2017
|
|
6CMR
 
 | | Closed structure of active SHP2 mutant E76D bound to SHP099 inhibitor | | Descriptor: | 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Padua, R.A.P, Sun, Y, Marko, I, Pitsawong, W, Kern, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2.
Nat Commun, 9, 2018
|
|
6CLF
 
 | | 1.15 A MicroED structure of GSNQNNF at 1.9 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLO
 
 | | 1.15 A MicroED structure of GSNQNNF at 2.1 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CMX
 
 | | Human Teneurin 2 extra-cellular region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Teneurin-2, alpha-D-mannopyranose, ... | | Authors: | Shalev-Benami, M, Li, J, Sudhof, T, Skiniotis, G, Arac, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-07-25 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for Teneurin Function in Circuit-Wiring: A Toxin Motif at the Synapse.
Cell, 173, 2018
|
|
9FBK
 
 | |
5JYA
 
 | |
6CJZ
 
 | |
6CBJ
 
 | | Crystal Structure of DH270.3 Fab in complex with Man9 | | Descriptor: | DH270.3 Fab heavy chain, DH270.3 Fab light chain, PHOSPHATE ION, ... | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | HIV envelope V3 region mimic embodies key features of a broadly neutralizing antibody lineage epitope.
Nat Commun, 9, 2018
|
|
9EMM
 
 | |
4WCE
 
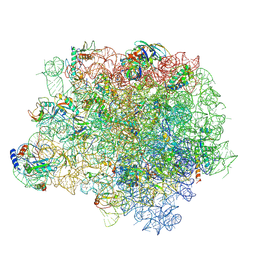 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S rRNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-09-04 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5K2D
 
 | | 1.9A angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
9EMN
 
 | |
5KHT
 
 | | Crystal structure of the N-terminal fragment of tropomyosin isoform Tpm1.1 at 1.5 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tropomyosin alpha-1 chain,General control protein GCN4 | | Authors: | Kostyukova, A.S, Krieger, I, Yoon, Y.-H, Tolkatchev, D, Samatey, F.A. | | Deposit date: | 2016-06-15 | | Release date: | 2017-06-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4964 Å) | | Cite: | Structural destabilization of tropomyosin induced by the cardiomyopathy-linked mutation R21H.
Protein Sci., 27, 2018
|
|
6CFS
 
 | |
