5FE1
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment BR004 (fragment 1) | | Descriptor: | 1,2-ETHANEDIOL, 1-METHYLQUINOLIN-2(1H)-ONE, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE8
 
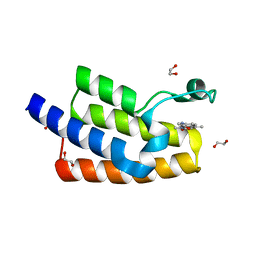 | | Crystal structure of human PCAF bromodomain in complex with compound SL1126 (compound 12) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Histone acetyltransferase KAT2B, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5EPK
 
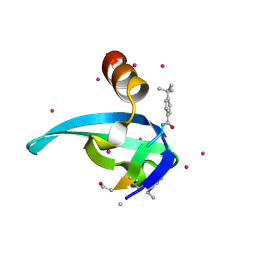 | | Crystal Structure of chromodomain of CBX2 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 2, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
4YY8
 
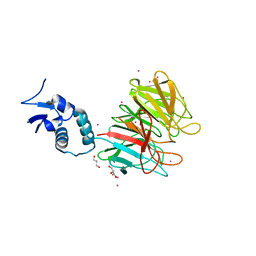 | | Crystal Structure Analysis of Kelch protein from Plasmodium falciparum | | Descriptor: | GLYCEROL, Kelch protein, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, El Bakkouri, M, Senisterra, G, Osman, K.T, Lovato, D.V, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure Analysis of Kelch protein from Plasmodium falciparum.
to be published
|
|
6OGN
 
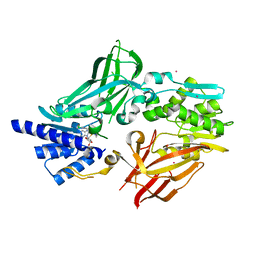 | | Crystal structure of mouse protein arginine methyltransferase 7 in complex with SGC8158 chemical probe | | Descriptor: | 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine, Protein arginine N-methyltransferase 7, UNKNOWN ATOM OR ION, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-03 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response.
Nat Commun, 11, 2020
|
|
5EK9
 
 | | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline inhibitor | | Descriptor: | Bromodomain-containing protein 2, propan-2-yl ~{N}-[(2~{S},4~{R})-1-ethanoyl-6-(furan-2-yl)-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Picaud, S, Fedorov, O, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploiting a water network to achieve enthalpy-driven, bromodomain-selective BET inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
5ELZ
 
 | | Staphylococcus aureus Type II pantothenate kinase in complex with a pantothenate analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, N-(1,3-benzodioxol-5-ylmethyl)-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Mottaghi, K, Hughes, S.J, Tempel, W, Hong, B, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-05 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent Pantothenamide Inhibitors of Staphylococcus aureus Pantothenate Kinase through a Minimal SAR Study: Inhibition Is Due to Trapping of the Product.
Acs Infect Dis., 2, 2016
|
|
5E8R
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | CHLORIDE ION, N-methyl-N-({4-[4-(propan-2-yloxy)phenyl]-1H-pyrrol-3-yl}methyl)ethane-1,2-diamine, Protein arginine N-methyltransferase 6, ... | | Authors: | DONG, A, ZENG, H, LIU, J, TEMPEL, W, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, JIN, J, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases.
Acs Chem.Biol., 11, 2016
|
|
5FE2
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment BR013 (fragment 3) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3~{H}-isoindol-1-one, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE9
 
 | | Crystal structure of human PCAF bromodomain in complex with compound SL1122 (compound 13) | | Descriptor: | 1,2-ETHANEDIOL, Histone acetyltransferase KAT2B, ~{N}-(1,4-dimethyl-2-oxidanylidene-quinolin-7-yl)methanesulfonamide | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5B8D
 
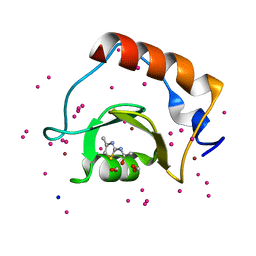 | | Crystal structure of a low occupancy fragment candidate (N-(4-Methyl-1,3-thiazol-2-yl)propanamide) bound adjacent to the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | FORMIC ACID, Histone deacetylase 6, SODIUM ION, ... | | Authors: | Harding, R.J, Tempel, W, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Ravichandran, M, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
3C90
 
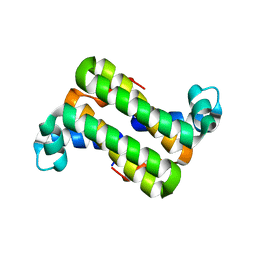 | | The 1.25 A Resolution Structure of Phosphoribosyl-ATP Pyrophosphohydrolase from Mycobacterium tuberculosis, crystal form II | | Descriptor: | Phosphoribosyl-ATP pyrophosphatase | | Authors: | Javid-Majd, F, Yang, D, Ioerger, T.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-02-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The 1.25 A resolution structure of phosphoribosyl-ATP pyrophosphohydrolase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7B33
 
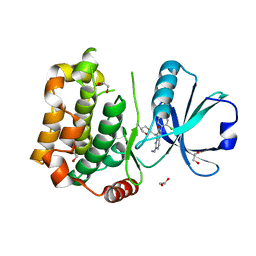 | | MST3 in complex with MRIA11 | | Descriptor: | 1,2-ETHANEDIOL, 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[4-[6-[bis(fluoranyl)methyl]pyridin-2-yl]-2-chloranyl-phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7B30
 
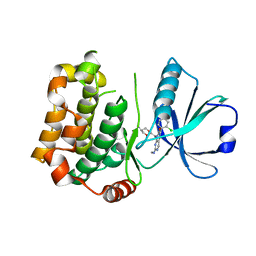 | | MST3 in complex with compound G-5555 | | Descriptor: | 8-[(trans-5-amino-1,3-dioxan-2-yl)methyl]-6-[2-chloro-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7B35
 
 | | MST3 in complex with compound MRIA13 | | Descriptor: | 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(3-methoxy-6-methyl-pyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.40005136 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7B31
 
 | | MST3 in complex with compound MRIA9 | | Descriptor: | 1-[(5-azanyl-1,3-dioxan-2-yl)methyl]-3-[2-chloranyl-4-(3-fluoranylpyridin-2-yl)phenyl]-7-(methylamino)-1,6-naphthyridin-2-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7B32
 
 | | MST3 in complex with MRIA7 | | Descriptor: | 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-[(2-methoxyphenyl)amino]pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7B34
 
 | | MST3 in complex with compound MRIA12 | | Descriptor: | 1,2-ETHANEDIOL, 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(3-fluoranyl-6-methyl-pyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7BE6
 
 | | Structure of DDR1 receptor tyrosine kinase in complex with inhibitor SR159 | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-N-(4-(((2S)-4-cyclohexyl-1-((1-(methylsulfonyl)piperidin-3-yl)amino)-1-oxobutan-2-yl)carbamoyl)benzyl)-1-phenyl-1H-pyrazole-4-carboxamide, Epithelial discoidin domain-containing receptor 1, ... | | Authors: | Pinkas, D.M, Bufton, J.C, Roehm, S, Joerger, A.C, Knapp, S, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87081933 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
7BE4
 
 | | Crystal structure of MAP kinase p38 alpha in complex with inhibitor SR159 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{R})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
6ZS2
 
 | | Crystal Structure of the bromodomain of human transcription activator BRG1 (SMARCA4) in complex with 2-(6-amino-5-(piperazin-1-yl)pyridazin-3-yl)phenol | | Descriptor: | 1,2-ETHANEDIOL, 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Transcription activator BRG1 | | Authors: | Preuss, F, Joerger, A.C, Kraemer, A, Wanior, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
6ZS4
 
 | | Crystal structure of the fifth bromodomain of human protein polybromo-1 in complex with tert-butyl 4-[3-amino-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazine-1-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, Protein polybromo-1, tert-butyl 4-[3-amino-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazine-1-carboxylate | | Authors: | Preuss, F, Joerger, A.C, Kraemer, A, Wanior, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
5C4Q
 
 | | Crystal Structure Analysis of bromodomain from Leishmania donovani complexed with bromosporine | | Descriptor: | Bromodomain, Bromosporine, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Amani, M, Hou, C.F.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-18 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Crystal Structure Analysis of bromodomain from Leishmania donovani complexed with bromosporine
to be published
|
|
7AWC
 
 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG)in complex with rosiglitazone | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), GLYCEROL, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Endogenous vitamin E metabolites mediate allosteric PPAR gamma activation with unprecedented co-regulatory interactions.
Cell Chem Biol, 28, 2021
|
|
7AWD
 
 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG)in complex with garcinoic acid | | Descriptor: | (2Z,6E,10E)-13-[(2R)-6-hydroxy-2,8-dimethyl-3,4-dihydro-2H-1-benzopyran-2-yl]-2,6,10-trimethyltrideca-2,6,10-trienoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Endogenous vitamin E metabolites mediate allosteric PPAR gamma activation with unprecedented co-regulatory interactions.
Cell Chem Biol, 28, 2021
|
|
