1V6I
 
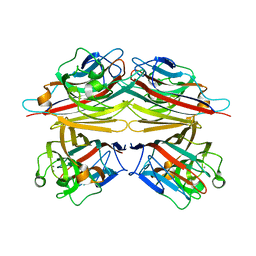 | | Peanut lectin-lactose complex in acidic pH | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Kundhavai Natchiar, S, Arockia Jeyaprakash, A, Ramya, T.N.C, Thomas, C.J, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural plasticity of peanut lectin: an X-ray analysis involving variation in pH, ligand binding and crystal structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3LIQ
 
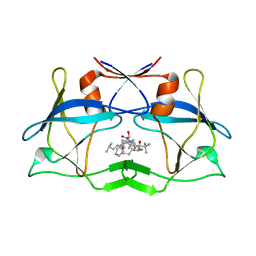 | | Crystal Structure of HTLV protease complexed with the inhibitor, KNI-10673 | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2S)-2-[[(2S)-2-azanyl-2-phenyl-ethanoyl]amino]-3,3-dimethyl-butanoyl]amino]-2-hydroxy-4-phenyl-but anoyl]-5,5-dimethyl-N-(2-methylpropyl)-1,3-thiazolidine-4-carboxamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
421P
 
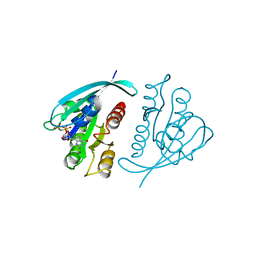 | | THREE-DIMENSIONAL STRUCTURES OF H-RAS P21 MUTANTS: MOLECULAR BASIS FOR THEIR INABILITY TO FUNCTION AS SIGNAL SWITCH MOLECULES | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Krengel, U, John, J, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules.
Cell(Cambridge,Mass.), 62, 1990
|
|
6RHZ
 
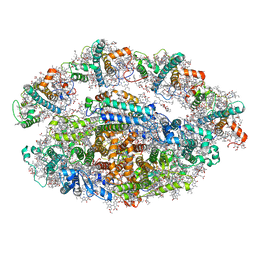 | | Structure of a minimal photosystem I from a green alga | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Perez Boerema, A, Klaiman, D, Caspy, I, Netzer-El, S.Y, Amunts, A, Nelson, N. | | Deposit date: | 2019-04-23 | | Release date: | 2020-02-19 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a minimal photosystem I from the green alga Dunaliella salina.
Nat.Plants, 6, 2020
|
|
3N9X
 
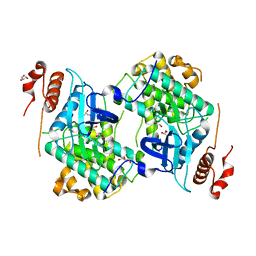 | | Crystal structure of Map Kinase from plasmodium berghei, PB000659.00.0 | | Descriptor: | GLYCEROL, Phosphotransferase | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, MacKenzie, F, Kozieradzki, I, Chau, I, Lew, J, Senisterra, G, Artz, J, Amani, M, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Map Kinase from plasmodium berghei, PB000659.00.0
To be Published
|
|
4O1N
 
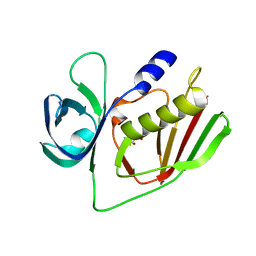 | |
5DIU
 
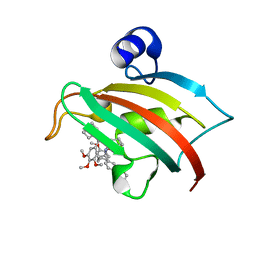 | | The Fk1 domain of FKBP51 in complex with the new synthetic ligand 2-(3-((R)-1-((S)-1-((S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)acetyl)piperidine-2-carboxamido)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-1-[({(2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)acetyl]piperidin-2-yl}carbonyl)amino]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetic acid | | Authors: | Gaali, S, Feng, X, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Rapid, Structure-Based Exploration of Pipecolic Acid Amides as Novel Selective Antagonists of the FK506-Binding Protein 51.
J.Med.Chem., 59, 2016
|
|
3LIY
 
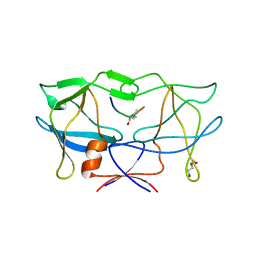 | | crystal structure of HTLV protease complexed with Statine-containing peptide inhibitor | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protease, statine-containing inhibitor | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
3NG6
 
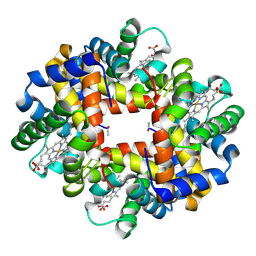 | | The crystal structure of hemoglobin I from Trematomus newnesi in deoxygenated state obtained through an oxidation/reduction cycle in which potassium hexacyanoferrate and sodium dithionite were alternatively added | | Descriptor: | Hemoglobin subunit alpha-1, Hemoglobin subunit beta-1/2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vergara, A, Vitagliano, L, Merlino, A, Sica, F, Marino, K, Mazzarella, L. | | Deposit date: | 2010-06-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An order-disorder transition plays a role in switching off the root effect in fish hemoglobins.
J.Biol.Chem., 285, 2010
|
|
2FE7
 
 | | The crystal structure of a probable N-acetyltransferase from Pseudomonas aeruginosa | | Descriptor: | probable N-acetyltransferase | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a N-acetyltransferase from Pseudomonas aeruginosa
To be Published
|
|
4H8J
 
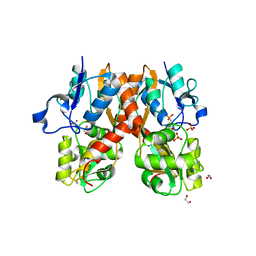 | | Structure of GluA2-LBD in complex with MES | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutamate receptor 2, ... | | Authors: | Reiter, A, Skerra, A, Trauner, D, Schiefner, A. | | Deposit date: | 2012-09-22 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of an artificial photoreceptor
To be Published
|
|
2EU1
 
 | | Crystal structure of the chaperonin GroEL-E461K | | Descriptor: | GROEL | | Authors: | Cabo-Bilbao, A, Spinelli, S, Sot, B, Agirre, J, Mechaly, A.E, Muga, A, Guerin, D.M.A. | | Deposit date: | 2005-10-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal structure of the temperature-sensitive and allosteric-defective chaperonin GroEL(E461K).
J.Struct.Biol., 155, 2006
|
|
2Q9N
 
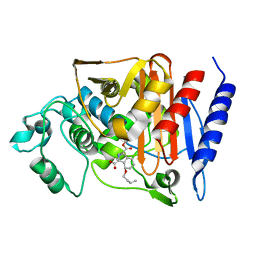 | | 4-Substituted Trinems as Broad Spectrum-Lactamase Inhibitors: Structure-based Design, Synthesis and Biological Activity | | Descriptor: | (1S,4R,7AR)-4-BUTOXY-1-[(1R)-1-FORMYLPROPYL]-2,4,5,6,7,7A-HEXAHYDRO-1H-ISOINDOLE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Plantan, I, Selic, L, Mesar, T, Stefanic Anderluh, P, Oblak, M, Prezelj, A, Hesse, L, Andrejasic, M, Vilar, M, Turk, D, Kocijan, A, Prevec, T, Vilfan, G, Kocjan, D, Copar, A, Urleb, U, Solmajer, T. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 4-Substituted Trinems as Broad Spectrum beta-Lactamase Inhibitors: Structure-Based Design, Synthesis, and Biological Activity
J.Med.Chem., 50, 2007
|
|
2FIW
 
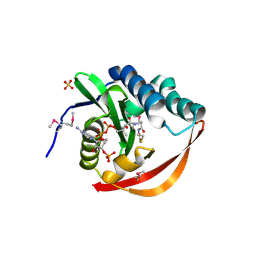 | | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris | | Descriptor: | ACETYL COENZYME *A, GCN5-related N-acetyltransferase:Aminotransferase, class-II, ... | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-30 | | Release date: | 2006-02-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris
To be Published, 2006
|
|
4H02
 
 | |
5HBH
 
 | | CDK8-CYCC IN COMPLEX WITH 5-{5-Chloro-4-[1-(2-methoxy-ethyl)-1,8-diaza-spiro[4.5]dec-8-yl]-pyridin-3-yl}-1-methyl-1,3-dihydro-benzo[c]isothiazole 2,2-dioxide | | Descriptor: | 1,2-ETHANEDIOL, 5-[5-chloranyl-4-[1-(2-methoxyethyl)-1,8-diazaspiro[4.5]decan-8-yl]pyridin-3-yl]-1-methyl-3~{H}-2,1-benzothiazole 2,2-dioxide, Cyclin-C, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Small-Molecule Modulators of the Mediator Complex-Associated Kinases CDK8 and CDK19.
J.Med.Chem., 59, 2016
|
|
3MTJ
 
 | | The Crystal Structure of a Homoserine Dehydrogenase from Thiobacillus denitrificans to 2.15A | | Descriptor: | Homoserine dehydrogenase, SULFATE ION | | Authors: | Stein, A.J, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-30 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of a Homoserine Dehydrogenase from Thiobacillus denitrificans to 2.15A
To be Published
|
|
2QET
 
 | |
3MV1
 
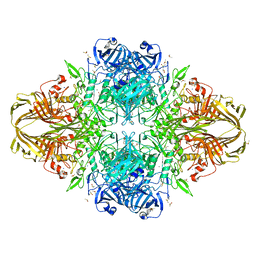 | | E.Coli (lacZ) beta-galactosidase (R599A) in complex with Guanidinium | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, GUANIDINE, ... | | Authors: | Dugdale, M.L, Vance, M, Driedger, M.L, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
5MJL
 
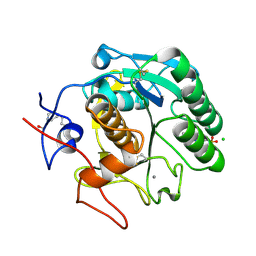 | | Single-shot pink beam serial crystallography: Proteinase K | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.21013784 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
2V2G
 
 | |
5G4I
 
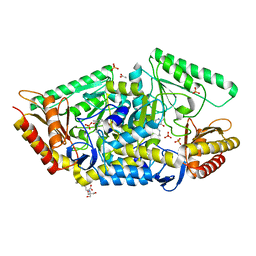 | | PLP-dependent phospholyase A1RDF1 from Arthrobacter aurescens TC1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Cuetos, A, Tuan, A.N, Mangas Sanchez, J, Grogan, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-19 | | Last modified: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Phospholyase Activity of a Class III Transaminase Homologue.
Chembiochem, 17, 2016
|
|
5B6W
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 16 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
3V75
 
 | | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680 | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Stogios, P.J, Xu, X, Cui, H, Kudritska, M, Tan, K, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680
To be Published
|
|
3BJH
 
 | | Soft-SAD crystal structure of a pheromone binding protein from the honeybee Apis mellifera L. | | Descriptor: | GLYCEROL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Lartigue, A, Gruez, A, Briand, L, Blon, F, Bezirard, V, Walsh, M, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sulfur single-wavelength anomalous diffraction crystal structure of a pheromone-binding protein from the honeybee Apis mellifera L.
J.Biol.Chem., 279, 2004
|
|
