7AVY
 
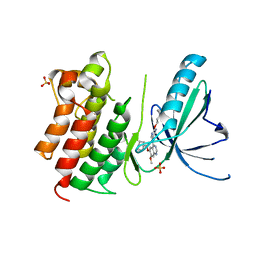 | | MerTK kinase domain in complex with quinazoline-based inhbitor | | Descriptor: | N-(2-(2-cyclopropylethoxy)pyrimidin-5-yl)-7-methoxy-6-(piperidin-4-ylmethoxy)quinazolin-4-amine, SULFATE ION, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AVX
 
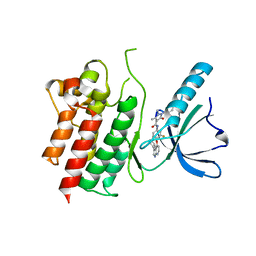 | | MerTK kinase domain in complex with NPS-1034 | | Descriptor: | 1-(4-fluorophenyl)-N-[3-fluoro-4-[(3-phenyl-1H-pyrrolo[2,3-b]pyridin-4-yl)oxy]phenyl]-2,3-dimethyl-5-oxopyrazole-4-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW2
 
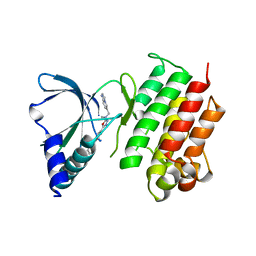 | | MerTK kinase domain with type 1.5 inhibitor from a DNA-encoded library | | Descriptor: | 5-(2'-chloro-[1,1'-biphenyl]-4-yl)-N-(imidazo[1,2-a]pyridin-6-ylmethyl)-N-methyl-1,3,4-oxadiazol-2-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
4HYU
 
 | |
7AW4
 
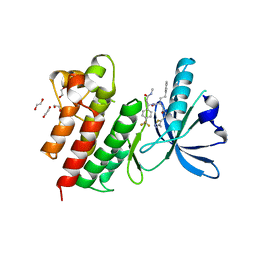 | | MerTK kinase domain with type 3 inhibitor from a DNA-encoded library | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-5-(4-methyl-1,2,3-thiadiazol-5-yl)-N-((R)-1-(((R)-3-(methylamino)-3-oxo-1-(4-(trifluoromethyl)phenyl)propyl)amino)-1-oxo-4-phenylbutan-2-yl)isoxazole-4-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
1P6X
 
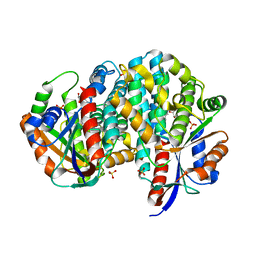 | | Crystal structure of EHV4-TK complexed with Thy and SO4 | | Descriptor: | SULFATE ION, THYMIDINE, Thymidine kinase | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
7AW1
 
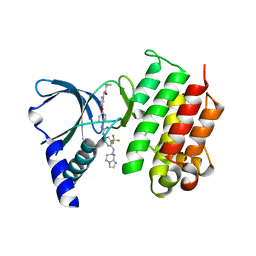 | | MerTK kinase domain in complex with a type 2 inhibitor | | Descriptor: | N-(6-(4-(3-(4-((5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)methyl)-3-(trifluoromethyl)phenyl)ureido)phenoxy)pyrimidin-4-yl)cyclopropanecarboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
3EDH
 
 | |
7AVZ
 
 | | MerTK kinase domain in complex with a bisaminopyrimidine inhibitor | | Descriptor: | (R)-N2-(4-(cyclopropylmethoxy)-3,5-difluorophenyl)-5-(3-methylpiperazin-1-yl)-N4-(tetrahydro-2H-pyran-4-yl)pyrimidine-2,4-diamine, Tyrosine-protein kinase Mer | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW3
 
 | | MerTK kinase domain with type 1 inhibitor from a DNA-encoded library | | Descriptor: | 2-(1-((5-chloro-1H-pyrrolo[2,3-b]pyridine-3-carboxamido)methyl)-2-azabicyclo[2.1.1]hexan-2-yl)-N-methyl-4-(trifluoromethyl)thiazole-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
1OW4
 
 | | Crystal structure of a pheromone binding protein from the cockroach Leucophaea maderae in complex with the fluorescent reporter ANS (1-anilinonaphtalene-8-sulfonic acid), | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, GLYCEROL, pheromone binding protein | | Authors: | Lartigue, A, Gruez, A, Spinelli, S, Riviere, S, Brossut, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-03-28 | | Release date: | 2003-08-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | THE CRYSTAL STRUCTURE OF A COCKROACH PHEROMONE-BINDING PROTEIN SUGGESTS A NEW LIGAND BINDING AND RELEASE MECHANISM
J.Biol.Chem., 278, 2003
|
|
1P9Q
 
 | | Structure of a hypothetical protein AF0491 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0491 | | Authors: | Savchenko, A, Evdokimova, E, Skarina, T, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
8GXX
 
 | | 3 nucleotide-bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
4XSN
 
 | | Copper(II) bound to the Z-DNA form of d(CGCGCG) | | Descriptor: | COPPER (II) ION, DNA (5'-D(*CP*(BGM)P*CP*GP*CP*GP)-3') | | Authors: | Rohner, M, Medina-Molner, A, Spingler, B. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | N,N,O and N,O,N Meridional cis Coordination of Two Guanines to Copper(II) by d(CGCGCG)2.
Inorg.Chem., 55, 2016
|
|
1WQU
 
 | | Solution structure of the human FES SH2 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase FES/FPS | | Authors: | Scott, A, Pantoja-Uceda, D, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Tanaka, A, Sugano, S, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-02 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Src homology 2 domain from the human feline sarcoma oncogene Fes
J.Biomol.NMR, 31, 2005
|
|
2JE5
 
 | | STRUCTURAL AND MECHANISTIC BASIS OF PENICILLIN BINDING PROTEIN INHIBITION BY LACTIVICINS | | Descriptor: | (2E)-2-{[(2S)-2-(ACETYLAMINO)-2-CARBOXYETHOXY]IMINO}PENTANEDIOIC ACID, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Macheboeuf, P, Fisher, D.S, Brown, T.J, Zervosen, A, Luxen, A, Joris, B, Dessen, A, Schofield, C.J. | | Deposit date: | 2007-01-15 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Basis of Penicillin-Binding Protein Inhibition by Lactivicins
Nat.Chem.Biol., 3, 2007
|
|
8GXY
 
 | | 2 sulfate-bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | SULFATE ION, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXU
 
 | | 1 ATP-bound V1EG of V/A-ATPase from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, SULFATE ION, V-type ATP synthase alpha chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXW
 
 | | 2 ATP-bound V1EG of V/A-ATPase from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXZ
 
 | | 1 sulfate and 1 ATP bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8G5E
 
 | | Crystal Structure of SETDB1 Tudor domain in complex with UNC6535 | | Descriptor: | Histone-lysine N-methyltransferase SETDB1, N~4~-[6-(dimethylamino)hexyl]-N~2~-[5-(dimethylamino)pentyl]-6,7-dimethoxyquinazoline-2,4-diamine, UNKNOWN ATOM OR ION | | Authors: | Beldar, S, Dong, A, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of SETDB1 Tudor domain in complex with UNC6535
To be published
|
|
5TF2
 
 | | CRYSTAL STRUCTURE OF THE WD40 DOMAIN OF THE HUMAN PROLACTIN REGULATORY ELEMENT-BINDING PROTEIN | | Descriptor: | Prolactin regulatory element-binding protein, UNKNOWN ATOM OR ION | | Authors: | Walker, J.R, Zhang, Q, Dong, A, Wernimont, A, Li, Y, He, H, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Chen, Z, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-23 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE OF THE WD40 DOMAIN OF THE HUMAN PROLACTIN REGULATORY ELEMENT-BINDING PROTEIN (CASP target)
To be published
|
|
8GCY
 
 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
8GS7
 
 | | SOLUTION NMR STRUCTURE OF N-TERMINAL DOMAIN OF TRICONEPHILA CLAVIPES MAJOR AMPULLATE SPIDROIN 2 | | Descriptor: | Major ampullate spidroin 2 variant 3 | | Authors: | Oktaviani, N.A, Malay, A.D, Matsugami, A, Hayashi, F, Numata, K. | | Deposit date: | 2022-09-05 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unusual p K a Values Mediate the Self-Assembly of Spider Dragline Silk Proteins.
Biomacromolecules, 24, 2023
|
|
8SBM
 
 | | Crystal structure of the wild-type Catalytic ATP-binding domain of Mtb DosS | | Descriptor: | 1,2-ETHANEDIOL, GAF domain-containing protein, SODIUM ION, ... | | Authors: | Larson, G, Shi, K, Aihara, H, Bhagi-Damodaran, A. | | Deposit date: | 2023-04-03 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Understanding ATP Binding to DosS Catalytic Domain with a Short ATP-Lid.
Biochemistry, 62, 2023
|
|
