5CCA
 
 | | Crystal structure of Mtb toxin | | Descriptor: | Endoribonuclease MazF3 | | Authors: | Cascio, D, Arbing, M, de Serrano, V, Eisenberg, D, Miallau, L, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Mtb toxin
To Be Published
|
|
5CCQ
 
 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | FORMIC ACID, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
1U16
 
 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) in complex with sulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Delta crystallin I, ... | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
5CE1
 
 | | Crystal Structure of Serine protease Hepsin in complex with Inhibitor | | Descriptor: | 2-[6-(1-hydroxycyclohexyl)pyridin-2-yl]-1H-indole-5-carboximidamide, Serine protease hepsin | | Authors: | Rao, K.N, Anita, R.C, Sangeetha, R, Anirudha, L, Subramnay, H. | | Deposit date: | 2015-07-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Serine protease Hepsin in complex with Inhibitor
To Be Published
|
|
7JVF
 
 | |
7K7A
 
 | | Transmembrane structure of TNFR1 | | Descriptor: | Tumor necrosis factor receptor superfamily member 1A | | Authors: | Zhao, L, Chou, J. | | Deposit date: | 2020-09-22 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Diversity and Similarity of Transmembrane Trimerization of TNF Receptors.
Front Cell Dev Biol, 8, 2020
|
|
6BNH
 
 | | Solution NMR structures of BRD4 ET domain with JMJD6 peptide | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, Bromodomain-containing protein 4 | | Authors: | Konuma, T, Yu, D, Zhao, C, Ju, Y, Sharma, R, Ren, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Sci Rep, 7, 2017
|
|
5CET
 
 | | Crystal structure of Rv2837c | | Descriptor: | Bifunctional oligoribonuclease and PAP phosphatase NrnA, MANGANESE (II) ION | | Authors: | Wang, F, He, Q, Zhu, D, Liu, S, Gu, L. | | Deposit date: | 2015-07-07 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Insight into the Mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP Phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
1U6F
 
 | | NMR solution structure of TcUBP1, a single RBD-unit from Trypanosoma cruzi | | Descriptor: | RNA-binding protein UBP1 | | Authors: | Volpon, L, D'orso, I, Frasch, A, Gehring, K. | | Deposit date: | 2004-07-29 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Study of TcUBP1, a Single RRM Domain Protein from Trypanosoma cruzi: Contribution of a beta Hairpin to RNA Binding
Biochemistry, 44, 2005
|
|
1X1Z
 
 | | Orotidine 5'-monophosphate decarboxylase (odcase) complexed with BMP (produced from 6-cyanoump) | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fujihashi, M, Bello, A.M, Poduch, E, Wei, L, Annedi, S.C, Pai, E.F, Kotra, L.P. | | Deposit date: | 2005-04-15 | | Release date: | 2005-12-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An unprecedented twist to ODCase catalytic activity
J.Am.Chem.Soc., 127, 2005
|
|
8OP2
 
 | |
5CO0
 
 | | Crystal Structure of the MTERF1 Y288A substitution bound to the termination sequence. | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3'), POTASSIUM ION, ... | | Authors: | Byrnes, J, Hauser, K, Norona, L, Mejia, E, Simmerling, C, Garcia-Diaz, M. | | Deposit date: | 2015-07-18 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Base Flipping by MTERF1 Can Accommodate Multiple Conformations and Occurs in a Stepwise Fashion.
J.Mol.Biol., 428, 2016
|
|
8OOU
 
 | |
5CN4
 
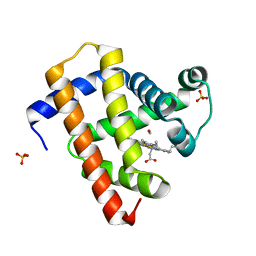 | | Ultrafast dynamics in myoglobin: -0.1 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
8OP1
 
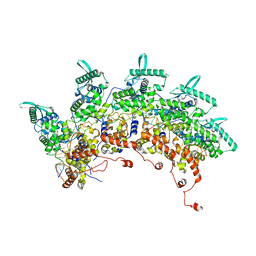 | | Subsection of a helical nucleocapsid of the Respiratory Syncytial Virus | | Descriptor: | Nucleoprotein, RNA (5'-R(P*CP*CP*CP*CP*CP*CP*C)-3') | | Authors: | Gonnin, L, Desfosses, A, Eleouet, J.F, Galloux, M, Gutsche, I. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural landscape of the respiratory syncytial virus nucleocapsids.
Nat Commun, 14, 2023
|
|
1WQD
 
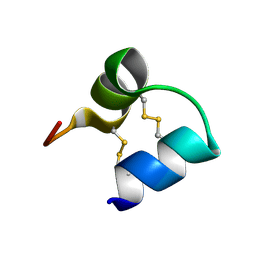 | | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis | | Descriptor: | OmTx2 | | Authors: | Chagot, B, Pimentel, C, Dai, L, Pil, J, Tytgat, J, Nakajima, T, Corzo, G, Darbon, H, Ferrat, G. | | Deposit date: | 2004-09-28 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis
Biochem.J., 388, 2005
|
|
1UFH
 
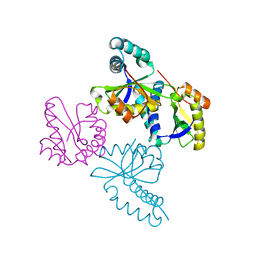 | | Structure of putative acetyltransferase, YYCN protein of Bacillus subtilis | | Descriptor: | YYCN protein | | Authors: | Taneja, B, Maar, S, Shuvalova, L, Collart, F.R, Anderson, W, Mondragon, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-29 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the bacillus subtilis YYCN protein: a putative N-acetyltransferase
Proteins, 53, 2003
|
|
8OIE
 
 | | Iron Nitrogenase Complex from Rhodobacter capsulatus | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Schmidt, F.V, Schulz, L, Zarzycki, J, Prinz, S, Erb, T.J, Rebelein, J.G. | | Deposit date: | 2023-03-22 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural insights into the iron nitrogenase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5CIU
 
 | | Structural basis of the recognition of H3K36me3 by DNMT3B PWWP domain | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, GLYCEROL, Histone H3.2 | | Authors: | Rondelet, G, DAL MASO, T, Willems, L, Wouters, J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B.
J.Struct.Biol., 194, 2016
|
|
1WT9
 
 | | crystal structure of Aa-X-bp-I, a snake venom protein with the activity of binding to coagulation factor X from Agkistrodon acutus | | Descriptor: | CALCIUM ION, agkisacutacin A chain, anticoagulant protein-B | | Authors: | Zhu, Z, Liu, S, Mo, X, Yu, X, Liang, Z, Zang, J, Zhao, W, Teng, M, Niu, L. | | Deposit date: | 2004-11-18 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterizations and Crystal structures of two snake venom proteins with the activity of binding coagulation factor X from Agkistrodon acutus
To be Published
|
|
6CG7
 
 | | mouse cadherin-22 EC1-2 adhesive fragment | | Descriptor: | CALCIUM ION, Cadherin-22 | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
6CGS
 
 | | mouse cadherin-7 EC1-2 adhesive fragment | | Descriptor: | CALCIUM ION, Cadherin-7, GLYCEROL | | Authors: | Brasch, J, Harrison, O.J, Kaczynska, A, Shapiro, L. | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
5CJX
 
 | |
1WYZ
 
 | | X-Ray structure of the putative methyltransferase from Bacteroides thetaiotaomicron VPI-5482 at the resolution 2.5 A. Norteast Structural Genomics Consortium target Btr28 | | Descriptor: | putative S-adenosylmethionine-dependent methyltransferase | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Vorobiev, S.M, Acton, T, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-21 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of the putative methyltransferase from Bacteroides thetaiotaomicron VPI-5482 at the resolution 2.5 A. Norteast Structural Genomics Consortium target Btr28
To be Published
|
|
5CNC
 
 | | Ultrafast dynamics in myoglobin: 0.6 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
