8KFX
 
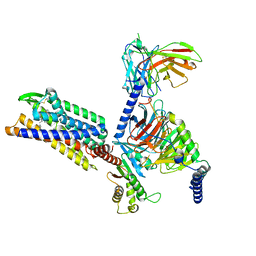 | | Gi bound CCR8 complex with nonpeptide agonist LMD-009 | | Descriptor: | 8-[[3-(2-methoxyphenoxy)phenyl]methyl]-1-(2-phenylethyl)-1,3,8-triazaspiro[4.5]decan-4-one, C-C chemokine receptor type 8,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
8KFZ
 
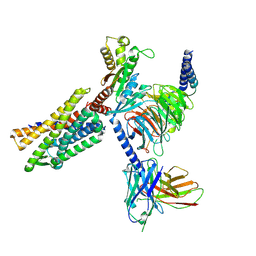 | | Gi bound CCR8 in ligand free state | | Descriptor: | C-C chemokine receptor type 8,LgBiT fusion protein,Recombinant Human Rhinovirus, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
8KFY
 
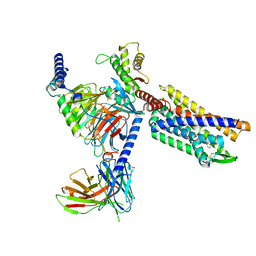 | | Gi bound CCR8 complex with nonpeptide agonist ZK 756326 | | Descriptor: | 2-[2-[4-[(3-phenoxyphenyl)methyl]piperazin-1-yl]ethoxy]ethanol, C-C chemokine receptor type 8,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
3H0H
 
 | |
7JMN
 
 | | Tail module of Mediator complex | | Descriptor: | MED15, Mediator of RNA polymerase II transcription subunit 14, Mediator of RNA polymerase II transcription subunit 16, ... | | Authors: | Zhang, H.Q, Chen, D.C. | | Deposit date: | 2020-08-02 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Mediator structure and conformation change.
Mol.Cell, 81, 2021
|
|
3PL9
 
 | | Crystal structure of spinach minor light-harvesting complex CP29 at 2.80 angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, M, Wan, T, Wang, L.F, Jia, C.J, Hou, Z.Q, Zhao, X.L, Zhang, J.P, Chang, W.R. | | Deposit date: | 2010-11-14 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into energy regulation of light-harvesting complex CP29 from spinach.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7KLZ
 
 | |
6Z45
 
 | | CDK9-Cyclin-T1 complex bound by compound 24 | | Descriptor: | (1~{S},3~{R})-3-acetamido-~{N}-[5-chloranyl-4-(5,5-dimethyl-4,6-dihydropyrrolo[1,2-b]pyrazol-3-yl)pyridin-2-yl]cyclohexane-1-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cyclin-T1, ... | | Authors: | Ferguson, A, Collie, G.W. | | Deposit date: | 2020-05-22 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Discovery of AZD4573, a Potent and Selective Inhibitor of CDK9 That Enables Short Duration of Target Engagement for the Treatment of Hematological Malignancies.
J.Med.Chem., 63, 2020
|
|
3H0I
 
 | |
6E9R
 
 | | DHF46 filament | | Descriptor: | DHF46 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
7XC3
 
 | |
7XC4
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain 3 (SARS-unique domain-M) in complex with Oxaprozin | | Descriptor: | 3-(4,5-diphenyl-1,3-oxazol-2-yl)propanoic acid, Papain-like protease nsp3 | | Authors: | Li, J, Liu, Y, Gao, J, Ruan, K. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Binding Sites of SARS-CoV-2 Macrodomain 3 Probed by Oxaprozin and Meclomen.
J.Med.Chem., 65, 2022
|
|
7DJZ
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW01 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, MW01 heavy chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
7DK0
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW05 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW05 heavy chain, MW05 light chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
5MQY
 
 | | CATHEPSIN L IN COMPLEX WITH 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile, Cathepsin L1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-12-21 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Prospective Evaluation of Free Energy Calculations for the Prioritization of Cathepsin L Inhibitors.
J. Med. Chem., 60, 2017
|
|
6P7Z
 
 | |
5MOM
 
 | |
7XWA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BA.4/5 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Suzuki, T, Kimura, K, Hashiguchi, T. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell, 185, 2022
|
|
4J6I
 
 | | Discovery of thiazolobenzoxepin PI3-kinase inhibitors that spare the PI3-kinase beta isoform | | Descriptor: | 2-methyl-1-(4-{2-[1-(2,2,2-trifluoroethyl)-1H-1,2,4-triazol-5-yl]-4,5-dihydro[1]benzoxepino[5,4-d][1,3]thiazol-8-yl}-1H-pyrazol-1-yl)propan-2-ol, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Rouge, L, Wu, P. | | Deposit date: | 2013-02-11 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of thiazolobenzoxepin PI3-kinase inhibitors that spare the PI3-kinase beta isoform.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6K32
 
 | | RdRp complex | | Descriptor: | 2'-O-methyladenosine 5'-(dihydrogen phosphate), 7-METHYLGUANOSINE, DIPHOSPHATE, ... | | Authors: | Li, X.W. | | Deposit date: | 2019-05-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of RdRps Within a Transcribing dsRNA Virus Provides Insights Into the Mechanisms of RNA Synthesis.
J.Mol.Biol., 432, 2020
|
|
7YOV
 
 | |
7YOT
 
 | |
7YOU
 
 | |
8HW4
 
 | | Cryo-EM structure of dehydroepiandrosterone sulfate-bound human ABC transporter ABCC3 in nanodiscs | | Descriptor: | 17-oxoandrost-5-en-3beta-yl hydrogen sulfate, ATP-binding cassette sub-family C member 3 | | Authors: | Wang, J, Wang, F.F, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2022-12-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket.
Embo J., 42, 2023
|
|
3MOW
 
 | | Crystal structure of SHP2 in complex with a tautomycetin analog TTN D-1 | | Descriptor: | (2Z)-2-[(1R)-3-{[(1R,2S,3R,6S,7S,10S,12S,15E,17E)-18-carboxy-16-ethyl-3,7-dihydroxy-1,2,6,10,12-pentamethyl-5-oxooctade ca-15,17-dien-1-yl]oxy}-1-hydroxy-3-oxopropyl]-3-methylbut-2-enedioic acid, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.-Y, Liu, S, Yu, Z, Yu, X. | | Deposit date: | 2010-04-23 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SHP2 in complex with a tautomycetin analog TTN D-1
To be Published
|
|
