4IK0
 
 | | Crystal structure of diaminopimelate epimerase Y268A mutant from Escherichia coli | | Descriptor: | Diaminopimelate epimerase, IODIDE ION | | Authors: | Hor, L, Dobson, R.C.J, Hutton, C.A, Perugini, M.A. | | Deposit date: | 2012-12-24 | | Release date: | 2013-02-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dimerization of bacterial diaminopimelate epimerase is essential for catalysis
J.Biol.Chem., 288, 2013
|
|
4IJZ
 
 | | Crystal structure of diaminopimelate epimerase from Escherichia coli | | Descriptor: | Diaminopimelate epimerase, NITRATE ION | | Authors: | Hor, L, Dobson, R.C.J, Hutton, C.A, Perugini, M.A. | | Deposit date: | 2012-12-24 | | Release date: | 2013-02-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dimerization of bacterial diaminopimelate epimerase is essential for catalysis
J.Biol.Chem., 288, 2013
|
|
3TUU
 
 | | Structure of dihydrodipicolinate synthase from the common grapevine | | Descriptor: | BROMIDE ION, CHLORIDE ION, dihydrodipicolinate synthase | | Authors: | Perugini, M.A, Dobson, R.C, Atkinson, S.C. | | Deposit date: | 2011-09-19 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal, Solution and In silico Structural Studies of Dihydrodipicolinate Synthase from the Common Grapevine.
Plos One, 7, 2012
|
|
3G05
 
 | | Crystal structure of N-terminal domain (2-550) of E.coli MnmG | | Descriptor: | SULFATE ION, tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-27 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
J.Bacteriol., 191, 2009
|
|
6GQE
 
 | |
3TXO
 
 | | PKC eta kinase in complex with a naphthyridine | | Descriptor: | 2-methyl-N~1~-[3-(pyridin-4-yl)-2,6-naphthyridin-1-yl]propane-1,2-diamine, Protein kinase C eta type | | Authors: | Stark, W, Rummel, G, Cowan-Jacob, S.W. | | Deposit date: | 2011-09-23 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2,6-Naphthyridines as potent and selective inhibitors of the novel protein kinase C isozymes.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5M96
 
 | |
2MI5
 
 | |
4PP8
 
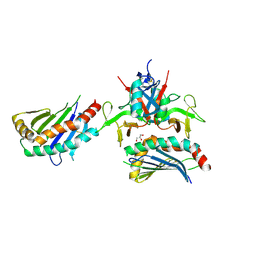 | |
2J3T
 
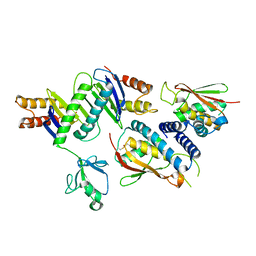 | | The crystal structure of the bet3-trs33-bet5-trs23 complex. | | Descriptor: | PALMITIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 1, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 3, ... | | Authors: | Kim, Y, Oh, B. | | Deposit date: | 2006-08-23 | | Release date: | 2006-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Architecture of the Multisubunit Trapp I Complex Suggests a Model for Vesicle Tethering.
Cell(Cambridge,Mass.), 127, 2006
|
|
1JFM
 
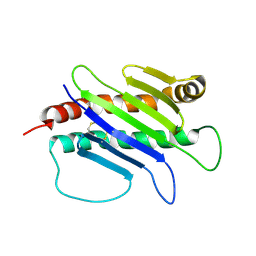 | |
3MZC
 
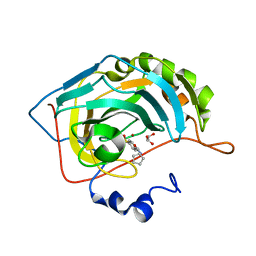 | | Human carbonic ahydrase II in complex with a benzenesulfonamide inhibitor | | Descriptor: | 4-[(cyclopentylcarbamoyl)amino]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, Mckenna, R. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Selective hydrophobic pocket binding observed within the carbonic anhydrase II active site accommodate different 4-substituted-ureido-benzenesulfonamides and correlate to inhibitor potency.
Chem.Commun.(Camb.), 46, 2010
|
|
3N2P
 
 | | Crystal structure of human carbonic anhydrase II in complex with a benzenesulfonamide inhibitor | | Descriptor: | 4-{[(3-nitrophenyl)carbamoyl]amino}benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, McKenna, R. | | Deposit date: | 2010-05-18 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Selective hydrophobic pocket binding observed within the carbonic anhydrase II active site accommodate different 4-substituted-ureido-benzenesulfonamides and correlate to inhibitor potency.
Chem.Commun.(Camb.), 46, 2010
|
|
3N3J
 
 | | Crystal structure of human carbonic anhydrase II in complex with a benzenesulfonamide inhibitor | | Descriptor: | 4-({[2-(1-methylethyl)phenyl]carbamoyl}amino)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, McKenna, R. | | Deposit date: | 2010-05-20 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective hydrophobic pocket binding observed within the carbonic anhydrase II active site accommodate different 4-substituted-ureido-benzenesulfonamides and correlate to inhibitor potency.
Chem.Commun.(Camb.), 46, 2010
|
|
3N4B
 
 | | Crystal structure of human carbonic anhydrase II in complex with a benzenesulfonamide inhibitor | | Descriptor: | 4-{[(4-fluorophenyl)carbamoyl]amino}benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, McKenna, R. | | Deposit date: | 2010-05-21 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective hydrophobic pocket binding observed within the carbonic anhydrase II active site accommodate different 4-substituted-ureido-benzenesulfonamides and correlate to inhibitor potency.
Chem.Commun.(Camb.), 46, 2010
|
|
3N0N
 
 | | Crystal structure of human carbonic anhydrase II in complex with a benzenesulfonamide inhibitor | | Descriptor: | 4-{[(pentafluorophenyl)carbamoyl]amino}benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, McKenna, R. | | Deposit date: | 2010-05-14 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective hydrophobic pocket binding observed within the carbonic anhydrase II active site accommodate different 4-substituted-ureido-benzenesulfonamides and correlate to inhibitor potency.
Chem.Commun.(Camb.), 46, 2010
|
|
