7CZS
 
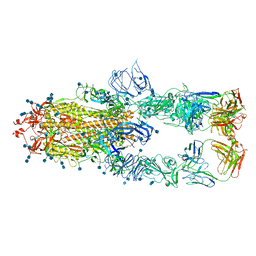 | | S protein of SARS-CoV-2 in complex bound with P5A-1B8_3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c542_heavy_IGHV3-53_IGHD3-10_IGHJ6,IGH@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZU
 
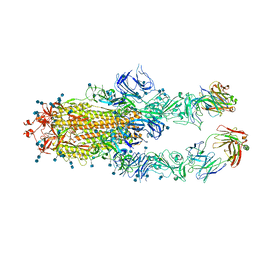 | | S protein of SARS-CoV-2 in complex bound with P5A-1B6_2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy variable 3-30-3,Immunoglobulin heavy variable 3-30-3,Chain H of P5A-1B6_2B,Immunoglobulin gamma-1 heavy chain,Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZV
 
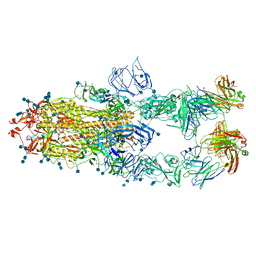 | | S protein of SARS-CoV-2 in complex bound with P5A-1B6_3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy variable 3-30-3,chain H of P5A-1B6_3B,Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZX
 
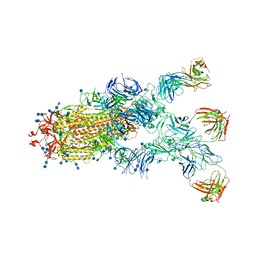 | | S protein of SARS-CoV-2 in complex bound with P5A-1B9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c168_light_IGKV4-1_IGKJ4,Uncharacterized protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZY
 
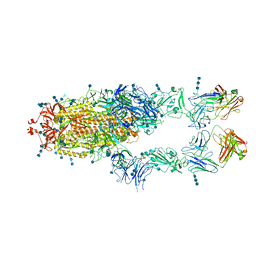 | | S protein of SARS-CoV-2 in complex bound with P5A-2F11_2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
5ZAM
 
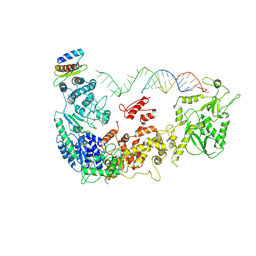 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2, RNA (73-mer) | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
5ZCS
 
 | | 4.9 Angstrom Cryo-EM structure of human mTOR complex 2 | | Descriptor: | Rapamycin-insensitive companion of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex 2 subunit MAPKAP1, ... | | Authors: | Chen, X, Liu, M, Tian, Y, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of human mTOR complex 2.
Cell Res., 28, 2018
|
|
3MQ7
 
 | | Crystal Structure of Ectodomain Mutant of BST-2/Tetherin/CD317 | | Descriptor: | Bone marrow stromal antigen 2, CALCIUM ION | | Authors: | Xiong, Y, Yang, H, Wang, J, Meng, W. | | Deposit date: | 2010-04-27 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insight into the mechanisms of enveloped virus tethering by tetherin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5ZAL
 
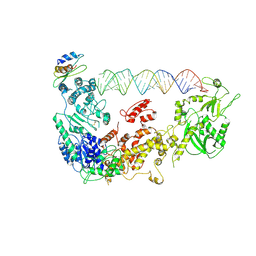 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2, RNA (73-mer) | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
5YZ0
 
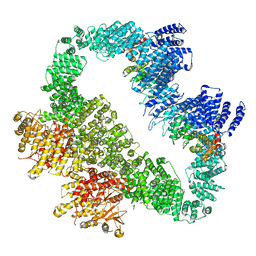 | | Cryo-EM Structure of human ATR-ATRIP complex | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR | | Authors: | Rao, Q, Liu, M, Tian, Y, Wu, Z, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of human ATR-ATRIP complex.
Cell Res., 28, 2018
|
|
5ZAK
 
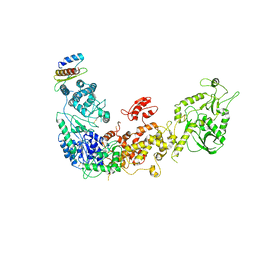 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
3MQB
 
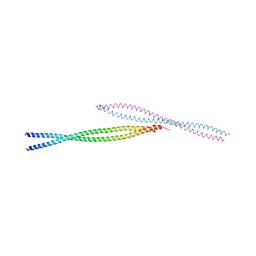 | |
3MQ9
 
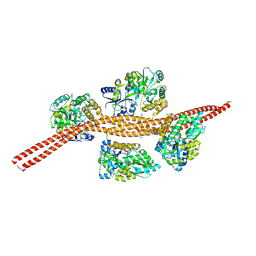 | |
3MQC
 
 | |
4M57
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 from maize | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10 | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
6VKL
 
 | | Negative stain reconstruction of the yeast exocyst octameric complex. | | Descriptor: | Exocyst complex component EXO70, Exocyst complex component EXO84, Exocyst complex component SEC10, ... | | Authors: | Frost, A, Munson, M. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Exocyst structural changes associated with activation of tethering downstream of Rho/Cdc42 GTPases.
J. Cell Biol., 219, 2020
|
|
4M59
 
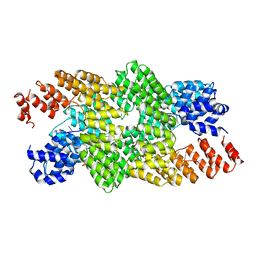 | | Crystal structure of the pentatricopeptide repeat protein PPR10 in complex with an 18-nt psaJ RNA element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
8YIH
 
 | | DmDcr-2/LoqsPD/slm1 in pre-dicing state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Cao, N, Su, S, Wang, J, Ma, J, Wang, H.-W. | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis of endo-siRNA processing by Drosophila Dicer-2 and Loqs-PD.
Nucleic Acids Res., 53, 2025
|
|
8YII
 
 | | DmDcr-2/LoqsPD/slm2 in dicing state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Cao, N, Su, S, Wang, J, Ma, J, Wang, H.-W. | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural basis of endo-siRNA processing by Drosophila Dicer-2 and Loqs-PD.
Nucleic Acids Res., 53, 2025
|
|
8YIG
 
 | | DmDcr-2/LoqsPD/slm2 in initial binding state | | Descriptor: | Dicer-2, isoform A, Isoform PD of Protein Loquacious, ... | | Authors: | Cao, N, Su, S, Wang, J, Ma, J, Wang, H.-W. | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of endo-siRNA processing by Drosophila Dicer-2 and Loqs-PD.
Nucleic Acids Res., 53, 2025
|
|
5G06
 
 | | Cryo-EM structure of yeast cytoplasmic exosome | | Descriptor: | EXOSOME COMPLEX COMPONENT CSL4, EXOSOME COMPLEX COMPONENT MTR3, EXOSOME COMPLEX COMPONENT RRP4, ... | | Authors: | Liu, J.J, Niu, C.Y, Wu, Y, Tan, D, Wang, Y, Ye, M.D, Liu, Y, Zhao, W.W, Zhou, K, Liu, Q.S, Dai, J.B, Yang, X.R, Dong, M.Q, Huang, N, Wang, H.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Cell Res., 26, 2016
|
|
5XMK
 
 | | Cryo-EM structure of the ATP-bound Vps4 mutant-E233Q complex with Vta1 (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
4ES4
 
 | | Crystal structure of YdiV and FlhD complex | | Descriptor: | Flagellar transcriptional regulator FlhD, Putative cyclic di-GMP regulator CdgR | | Authors: | Li, B, Gu, L. | | Deposit date: | 2012-04-22 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
3TLQ
 
 | | Crystal structure of EAL-like domain protein YdiV | | Descriptor: | GLYCEROL, PHOSPHATE ION, Regulatory protein YdiV | | Authors: | Li, B, Li, N, Wang, F, Guo, L, Liu, C, Zhu, D, Xu, S, Gu, L. | | Deposit date: | 2011-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
7CZZ
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-2F11_3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
