2YJ1
 
 | | Puma BH3 foldamer in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-PUMA BH3 FOLDAMER, BCL-2-LIKE PROTEIN 1 | | Authors: | Lee, E.F, Smith, B.J, Horne, W.S, Mayer, K.N, Evangelista, M, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-05-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Basis of Bcl-Xl Recognition by a Bh3-Mimetic Alpha-Beta-Peptide Generated Via Sequence-Based Design
Chembiochem, 12, 2011
|
|
3CB3
 
 | | Crystal structure of L-Talarate dehydratase from Polaromonas sp. JS666 complexed with Mg and L-glucarate | | Descriptor: | L-GLUCARIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of L-Talarate dehydratase from Polaromonas sp. JS666 complexed with Mg and L-glucarate.
To be Published
|
|
3C3F
 
 | |
3S84
 
 | | Dimeric apoA-IV | | Descriptor: | Apolipoprotein A-IV, SULFATE ION | | Authors: | Deng, X, Davidson, W.S, Thompson, T.B. | | Deposit date: | 2011-05-27 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Dimeric Apolipoprotein A-IV and Its Mechanism of Self-Association.
Structure, 20, 2012
|
|
7W1F
 
 | | Crystal structure of the dNTP triphosphohydrolase PA1124 from Pseudomonas aeruginosa | | Descriptor: | NICKEL (II) ION, Probable deoxyguanosinetriphosphate triphosphohydrolase | | Authors: | Oh, H.B, Song, W.S, Lee, K.C, Park, S.C, Yoon, S.I. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the dNTP triphosphohydrolase PA1124 from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 589, 2022
|
|
7WKV
 
 | | Crystal structure of human ALKBH5 in complex with 2-oxoglutarate (2OG) and m6A-containing ssRNA | | Descriptor: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, RNA (5'-R(P*GP*GP*(6MZ)P*C)-3'), ... | | Authors: | Kaur, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | Deposit date: | 2022-01-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanisms of substrate recognition and N6-methyladenosine demethylation revealed by crystal structures of ALKBH5-RNA complexes.
Nucleic Acids Res., 50, 2022
|
|
7WL0
 
 | | Crystal structure of human ALKBH5 in complex with N-oxalylglycine (NOG) and m6A-containing ssRNA | | Descriptor: | FORMIC ACID, MANGANESE (II) ION, N-OXALYLGLYCINE, ... | | Authors: | Kaur, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms of substrate recognition and N6-methyladenosine demethylation revealed by crystal structures of ALKBH5-RNA complexes.
Nucleic Acids Res., 50, 2022
|
|
7XFP
 
 | |
7X9R
 
 | | Crystal structure of the antirepressor GmaR | | Descriptor: | Glycosyl transferase family 2 | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
7X9S
 
 | | Crystal structure of a complex between the antirepressor GmaR and the transcriptional repressor MogR | | Descriptor: | GmaR, Motility gene repressor MogR | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
7W9Z
 
 | | Crystal structure of Bacillus subtilis YugJ in complex with NADP and nitrate | | Descriptor: | Iron-containing alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NITRATE ION | | Authors: | Cho, H.Y, Nam, M.S, Hong, H.J, Song, W.S, Yoon, S.I. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis.
Int J Mol Sci, 23, 2022
|
|
7W9Y
 
 | | Crystal structure of Bacillus subtilis YugJ in complex with NADP and nickel | | Descriptor: | Iron-containing alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICKEL (II) ION | | Authors: | Cho, H.Y, Nam, M.S, Hong, H.J, Song, W.S, Yoon, S.I. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis.
Int J Mol Sci, 23, 2022
|
|
7W9X
 
 | | Crystal structure of Bacillus subtilis YugJ in complex with nickel | | Descriptor: | Iron-containing alcohol dehydrogenase, NICKEL (II) ION | | Authors: | Cho, H.Y, Nam, M.S, Hong, H.J, Song, W.S, Yoon, S.I. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis.
Int J Mol Sci, 23, 2022
|
|
7XAY
 
 | | Crystal structure of Hat1-Hat2-Asf1-H3-H4 | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yue, Y, Yang, W.S, Xu, R.M. | | Deposit date: | 2022-03-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Topography of histone H3-H4 interaction with the Hat1-Hat2 acetyltransferase complex.
Genes Dev., 36, 2022
|
|
6H2B
 
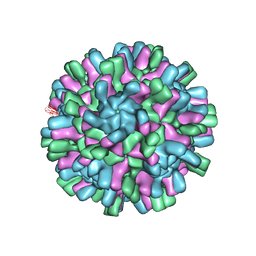 | | Structure of the Macrobrachium rosenbergii Nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Ho, K.H, Gabrielsen, M, Beh, P.L, Kueh, C.L, Thong, Q.X, Streetley, J, Tan, W.S, Bhella, D. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae?
PLoS Biol., 16, 2018
|
|
5BMH
 
 | | Nitroxide Spin Labels in Protein GB1: T44 Mutant, Crystal Form B | | Descriptor: | Immunoglobulin G-binding protein G, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Cunningham, T.C, Horne, W.S, Saxena, S. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rotameric preferences of a protein spin label at edge-strand beta-sheet sites.
Protein Sci., 25, 2016
|
|
5BMG
 
 | | Nitroxide Spin Labels in Protein GB1: E15 Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Immunoglobulin G-binding protein G, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Cunningham, T.C, Horne, W.S, Saxena, S. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rotameric preferences of a protein spin label at edge-strand beta-sheet sites.
Protein Sci., 25, 2016
|
|
7FHE
 
 | | Structure of prenyltransferase mutant Q295F from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHC
 
 | | Structure of prenyltransferase mutant V49W from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHB
 
 | | Structure of prenyltransferase from Streptomyces sp. (strain CL190) with bound GPP | | Descriptor: | GERANYL DIPHOSPHATE, Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHD
 
 | | Structure of prenyltransferase mutant Y288P from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHF
 
 | | Structure of prenyltransferase mutant V49W/Y288F/Q295F from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
3ZXJ
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HIAXHD3, ... | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXK
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIAXHD3, alpha-L-arabinofuranose-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXL
 
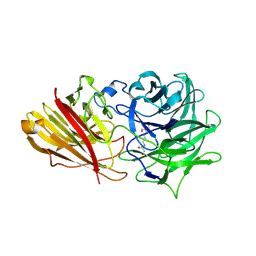 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HIAXHD3 | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
