2VWB
 
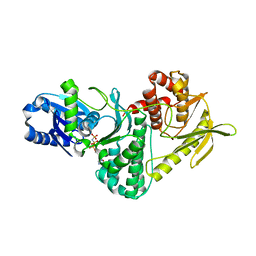 | | Structure of the archaeal Kae1-Bud32 fusion protein MJ1130: a model for the eukaryotic EKC-KEOPS subcomplex involved in transcription and telomere homeostasis. | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PUTATIVE O-SIALOGLYCOPROTEIN ENDOPEPTIDASE | | Authors: | Hecker, A, Lopreiato, R, Graille, M, Collinet, B, Forterre, P, Domenico, L, van Tilbeurgh, H. | | Deposit date: | 2008-06-20 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of the Archaeal Kae1/Bud32 Fusion Protein Mj1130: A Model for the Eukaryotic Ekc/Keops Subcomplex
Embo J., 27, 2008
|
|
2W9M
 
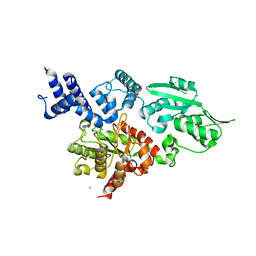 | | Structure of family X DNA polymerase from Deinococcus radiodurans | | Descriptor: | MERCURY (II) ION, POLYMERASE X, ZINC ION | | Authors: | Leulliot, N, Cladiere, L, Lecointe, F, Durand, D, Hubscher, U, van Tilbeurgh, H. | | Deposit date: | 2009-01-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The Family X DNA Polymerase from Deinococcus Radioduran Adopts a Non-Standard Extended Conformation.
J.Biol.Chem., 284, 2009
|
|
2VGM
 
 | |
2VGN
 
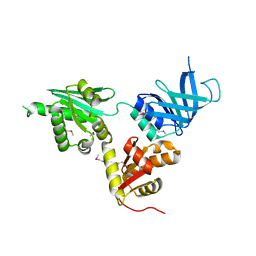 | | Structure of S. cerevisiae Dom34, a translation termination-like factor involved in RNA quality control pathways and interacting with Hbs1 (SelenoMet-labeled protein) | | Descriptor: | DOM34, GLYCEROL, PHOSPHATE ION | | Authors: | Graille, M, Chaillet, M, Van Tilbeurgh, H. | | Deposit date: | 2007-11-14 | | Release date: | 2008-01-22 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Structure of Yeast Dom34: A Protein Related to Translation Termination Factor Erf1 and Involved in No-Go Decay.
J.Biol.Chem., 283, 2008
|
|
3LTM
 
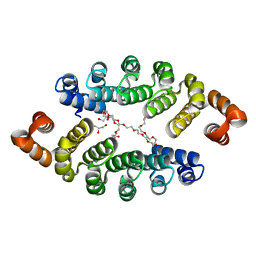 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | Descriptor: | Alpha-Rep4, DODECAETHYLENE GLYCOL, GLYCEROL, ... | | Authors: | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | Deposit date: | 2010-02-16 | | Release date: | 2010-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
4HV0
 
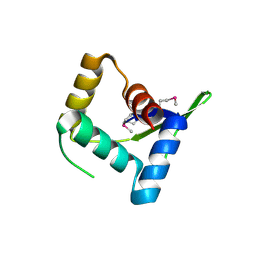 | | Structure and Function of AvtR, a Novel Transcriptional Regulator from a Hyperthermophilic Archaeal Lipothrixvirus | | Descriptor: | AvtR | | Authors: | Peixeiro, N, Keller, J, Collinet, B, Leulliot, N, Campanacci, V, Cortez, D, Cambillau, C, Nitta, K.R, Vincentelli, R, Forterre, P, Prangishvili, D, Sezonov, G, van Tilbeurgh, H. | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-21 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of AvtR, a Novel Transcriptional Regulator from a Hyperthermophilic Archaeal Lipothrixvirus.
J.Virol., 87, 2013
|
|
4JW3
 
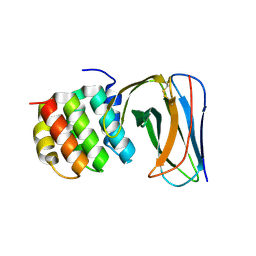 | | Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep) | | Descriptor: | Alpha-helical artificial proteins, Neocarzinostatin | | Authors: | Guellouz, A, Valerio-Lepiniec, M, Urvoas, A, Chevrel, A, Graille, M, Fourati-Kammoun, Z, Desmadril, M, van Tilbeurgh, H, Minard, P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selection of Specific Protein Binders for Pre-Defined Targets from an Optimized Library of Artificial Helicoidal Repeat Proteins (alphaRep).
Plos One, 8, 2013
|
|
4H18
 
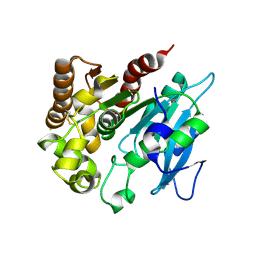 | | Three dimensional structure of corynomycoloyl tranferase C | | Descriptor: | Cmt1, MAGNESIUM ION | | Authors: | Huc, E, de Sousa D'Auria, C, Li de la Sierra-Gallay, I, Salmeron, C.H, van Tilbeurgh, H, Bayan, N, Houssin, C.H, Daffe, M, Tropis, M. | | Deposit date: | 2012-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Identification of a mycoloyl transferase selectively involved in o-acylation of polypeptides in corynebacteriales.
J.Bacteriol., 195, 2013
|
|
4JW2
 
 | | Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep) | | Descriptor: | 1,2-ETHANEDIOL, A3 artificial protein, bA3-2: binder of A3 protein | | Authors: | Guellouz, A, Valerio-Lepiniec, M, Urvoas, A, Chevrel, A, Graille, M, Fourati-Kammoun, Z, Desmadril, M, van Tilbeurgh, H, Minard, P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selection of Specific Protein Binders for Pre-Defined Targets from an Optimized Library of Artificial Helicoidal Repeat Proteins (alphaRep).
Plos One, 8, 2013
|
|
3QTM
 
 | | Structure of S. pombe nuclear import adaptor Nro1 (Space group P21) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Rispal, D, Henri, J, van Tilbeurgh, H, Graille, M, Seraphin, B. | | Deposit date: | 2011-02-23 | | Release date: | 2011-09-21 | | Last modified: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional analysis of Nro1/Ett1: a protein involved in translation termination in S. cerevisiae and in O2-mediated gene control in S. pombe
Rna, 17, 2011
|
|
3QTN
 
 | | Structure of S. pombe nuclear import adaptor Nro1 (Space group P6522) | | Descriptor: | Uncharacterized protein C4B3.07 | | Authors: | Rispal, D, Henri, J, van Tilbeurgh, H, Graille, M, Seraphin, B. | | Deposit date: | 2011-02-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.499 Å) | | Cite: | Structural and functional analysis of Nro1/Ett1: a protein involved in translation termination in S. cerevisiae and in O2-mediated gene control in S. pombe
Rna, 17, 2011
|
|
4FPR
 
 | | Structure of a fungal protein | | Descriptor: | Avirulence Effector AvrLm4-7 | | Authors: | Blondeau, K, Blaise, F, Graille, M, Linglin, J, Ollivier, B, Labarde, A, Doizy, A, Daverdin, G, Balesdent, M.H, Rouxel, T, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2012-06-22 | | Release date: | 2013-12-11 | | Last modified: | 2022-08-03 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structure of the effector AvrLm4-7 of Leptosphaeria maculans reveals insights into its translocation into plant cells and recognition by resistance proteins.
Plant J., 83, 2015
|
|
4FPQ
 
 | | Structure of a fungal protein | | Descriptor: | Avirulence Effector AvrLm4-7 | | Authors: | Blondeau, K, Blaise, F, Graille, M, Linglin, J, Ollivier, B, Labarde, A, Doizy, A, Daverdin, G, Balesdent, MH, Rouxel, T, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2012-06-22 | | Release date: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the effector AvrLm4-7 of Leptosphaeria maculans reveals insights into its translocation into plant cells and recognition by resistance proteins.
Plant J., 83, 2015
|
|
3DJW
 
 | | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1 | | Descriptor: | ORF99 | | Authors: | Goulet, A, Spinelli, S, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1
Protein Sci., 18, 2009
|
|
3DF6
 
 | | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1 | | Descriptor: | CALCIUM ION, ORF99 | | Authors: | Goulet, A, Spinelli, S, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-06-11 | | Release date: | 2009-06-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1
Protein Sci., 18, 2009
|
|
3MGU
 
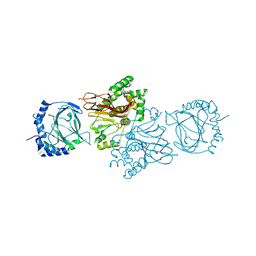 | | Structure of S. cerevisiae Tpa1 protein, a proline hydroxylase modifying ribosomal protein Rps23 | | Descriptor: | FE (II) ION, PKHD-type hydroxylase TPA1 | | Authors: | Henri, J, Rispal, D, Bayart, E, van Tilbeurgh, H, Seraphin, B, Graille, M. | | Deposit date: | 2010-04-07 | | Release date: | 2010-07-14 | | Last modified: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional insights into S. cerevisiae Tpa1, a putative prolyl hydroxylase influencing translation termination and transcription
To be Published
|
|
2J8A
 
 | | X-ray structure of the N-terminus RRM domain of Set1 | | Descriptor: | HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC | | Authors: | Tresaugues, L, Dehe, P.M, Guerois, R, Rodriguez-Gil, A, Varlet, I, Salah, P, Pamblanco, M, Luciano, P, Quevillon-Cheruel, S, Sollier, J, Leulliot, N, Couprie, J, Tordera, V, Zinn-Justin, S, Chavez, S, Van Tilbeurgh, H, Geli, V. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Structure of the N-Terminus Rrm Domain of Set1
To be Published
|
|
3M7P
 
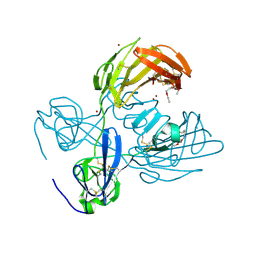 | | Fibronectin fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DODECAETHYLENE GLYCOL, FN1 protein, ... | | Authors: | Graille, M, Pagano, M, Rose, T, Reboud Ravaux, M, van Tilbeurgh, H. | | Deposit date: | 2010-03-17 | | Release date: | 2010-06-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Zinc Induces Structural Reorganization of Gelatin Binding Domain from Human Fibronectin and Affects Collagen Binding
Structure, 18, 2010
|
|
3Q87
 
 | | Structure of E. cuniculi Mtq2-Trm112 complex responible for the methylation of eRF1 translation termination factor | | Descriptor: | N6 adenine specific DNA methylase, Putative uncharacterized protein ECU08_1170, S-ADENOSYLMETHIONINE, ... | | Authors: | Liger, D, Mora, L, Lazar, N, Figaro, S, Henri, J, Scrima, N, Buckingham, R.H, van Tilbeurgh, H, Heurgue-Hamard, V, Graille, M. | | Deposit date: | 2011-01-06 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Mechanism of activation of methyltransferases involved in translation by the Trm112 'hub' protein
Nucleic Acids Res., 2011
|
|
4LUN
 
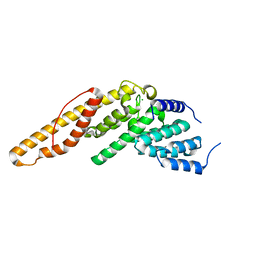 | | Structure of the N-terminal mIF4G domain from S. cerevisiae Upf2, a protein involved in the degradation of mRNAs containing premature stop codons | | Descriptor: | CHLORIDE ION, Nonsense-mediated mRNA decay protein 2 | | Authors: | Fourati, Z, Roy, B, Millan, C, Courreux, P.D, Kervestin, S, van Tilbeurgh, H, He, F, Uson, I, Jacobson, A, Graille, M. | | Deposit date: | 2013-07-25 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | A highly conserved region essential for NMD in the Upf2 N-terminal domain.
J.Mol.Biol., 426, 2014
|
|
4ML3
 
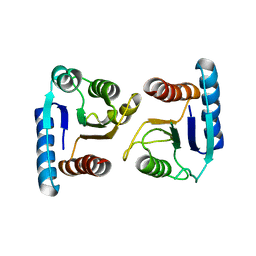 | | X-ray structure of ComE D58A REC domain from Streptococcus pneumoniae | | Descriptor: | Response regulator | | Authors: | Boudes, M, Sanchez, D, Durand, D, Graille, M, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2013-09-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae.
Nucleic Acids Res., 42, 2014
|
|
4MLD
 
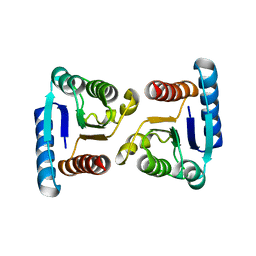 | | X-ray structure of ComE D58E REC domain from Streptococcus pneumoniae | | Descriptor: | Response regulator | | Authors: | Boudes, M, Sanchez, D, Durand, D, Graille, M, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2013-09-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae.
Nucleic Acids Res., 42, 2014
|
|
4P78
 
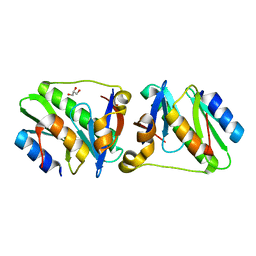 | | HicA3 and HicB3 toxin-antitoxin complex | | Descriptor: | GLYCEROL, HicA3 Toxin, HicB3 antitoxin | | Authors: | Li de la Sierra-Gallay, I, Bibi-Triki, S, van Tilbeurgh, H, Lazar, N, Pradel, E. | | Deposit date: | 2014-03-26 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Functional and Structural Analysis of HicA3-HicB3, a Novel Toxin-Antitoxin System of Yersinia pestis.
J.Bacteriol., 196, 2014
|
|
4P7D
 
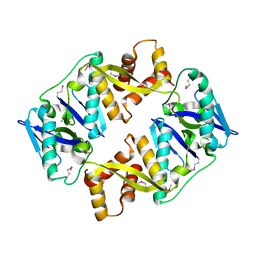 | | Antitoxin HicB3 crystal structure | | Descriptor: | Antitoxin HicB3, CHLORIDE ION | | Authors: | Li de la Sierra-Gallay, I, Bibi-Triki, S, van Tilbeurgh, H, Lazar, N, Pradel, E. | | Deposit date: | 2014-03-27 | | Release date: | 2014-08-27 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Functional and Structural Analysis of HicA3-HicB3, a Novel Toxin-Antitoxin System of Yersinia pestis.
J.Bacteriol., 196, 2014
|
|
2CFA
 
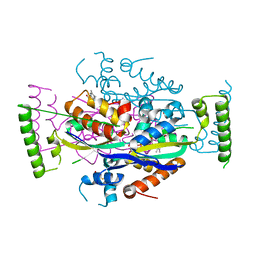 | | Structure of viral flavin-dependant thymidylate synthase ThyX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, THYMIDYLATE SYNTHASE | | Authors: | Graziani, S, Bernauer, J, Skouloubris, S, Graille, M, Zhou, C.-Z, Marchand, C, Decottignies, P, van Tilbeurgh, H, Myllykallio, H, Liebl, U. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic Mechanism and Structure of Viral Flavin-Dependent Thymidylate Synthase Thyx.
J.Biol.Chem., 281, 2006
|
|
